class: left background-image: url(https://images.unsplash.com/photo-1528830639624-fc16697bb8e7?q=80&w=1470&auto=format&fit=crop&ixlib=rb-4.0.3&ixid=M3wxMjA3fDB8MHxwaG90by1wYWdlfHx8fGVufDB8fHx8fA%3D%3D) background-size: cover <br> <br> <br> # <span style="color:burlywood">faceting in ggplot2 with reference data layer</span> ### <span style="color:burlywood">.tiny[Gina Reynolds | 2024-02-07 | Image credit: Georgi Petrov, Upsplash]</span> --- ### <span style="color:burlywood">Small multiples (facets) are a powerful tool.</span> -- ### <span style="color:burlywood">They break up and display data by category, so patterns within each category can be appreciated.</span> -- ### <span style="color:burlywood">But a simple facet means that comparison to the whole becomes more of an exercise in mental gymnastics.</span> -- ### <span style="color:burlywood">A solution is to present a reference layer of all the data beneath the featured category data. Let's look at the syntax in ggplot2.</span> --- count: false .panel1-feature-auto[ ```r *library(tidyverse) ``` ] .panel2-feature-auto[ ] --- count: false .panel1-feature-auto[ ```r library(tidyverse) *palmerpenguins::penguins ``` ] .panel2-feature-auto[ ``` ## # A tibble: 344 × 8 ## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g ## <fct> <fct> <dbl> <dbl> <int> <int> ## 1 Adelie Torgersen 39.1 18.7 181 3750 ## 2 Adelie Torgersen 39.5 17.4 186 3800 ## 3 Adelie Torgersen 40.3 18 195 3250 ## 4 Adelie Torgersen NA NA NA NA ## 5 Adelie Torgersen 36.7 19.3 193 3450 ## 6 Adelie Torgersen 39.3 20.6 190 3650 ## 7 Adelie Torgersen 38.9 17.8 181 3625 ## 8 Adelie Torgersen 39.2 19.6 195 4675 ## 9 Adelie Torgersen 34.1 18.1 193 3475 ## 10 Adelie Torgersen 42 20.2 190 4250 ## # ℹ 334 more rows ## # ℹ 2 more variables: sex <fct>, year <int> ``` ] --- count: false .panel1-feature-auto[ ```r library(tidyverse) palmerpenguins::penguins |> * ggplot(aes(x = bill_length_mm, y = bill_depth_mm)) ``` ] .panel2-feature-auto[ <!-- --> ] --- count: false .panel1-feature-auto[ ```r library(tidyverse) palmerpenguins::penguins |> ggplot(aes(x = bill_length_mm, y = bill_depth_mm)) + * facet_wrap(~ species) ``` ] .panel2-feature-auto[ <!-- --> ] --- count: false .panel1-feature-auto[ ```r library(tidyverse) palmerpenguins::penguins |> ggplot(aes(x = bill_length_mm, y = bill_depth_mm)) + facet_wrap(~ species) + * ggstamp::theme_void_fill("linen") ``` ] .panel2-feature-auto[ <!-- --> ] --- count: false .panel1-feature-auto[ ```r library(tidyverse) palmerpenguins::penguins |> ggplot(aes(x = bill_length_mm, y = bill_depth_mm)) + facet_wrap(~ species) + ggstamp::theme_void_fill("linen") + # plot all data in each species facet * geom_point(data = * . %>% select(-species), * size = 3, shape = 21, stroke = .7, * color = "burlywood4", fill= "white", * alpha = .7) ``` ] .panel2-feature-auto[ 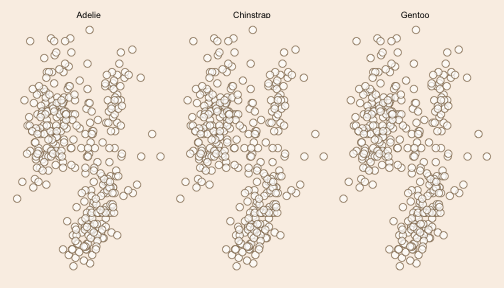<!-- --> ] --- count: false .panel1-feature-auto[ ```r library(tidyverse) palmerpenguins::penguins |> ggplot(aes(x = bill_length_mm, y = bill_depth_mm)) + facet_wrap(~ species) + ggstamp::theme_void_fill("linen") + # plot all data in each species facet geom_point(data = . %>% select(-species), size = 3, shape = 21, stroke = .7, color = "burlywood4", fill= "white", alpha = .7) + # use global data and plot each species in its facet * geom_point(size = 3, shape = 21, stroke = .2, * color = "white", fill = "burlywood4") ``` ] .panel2-feature-auto[ 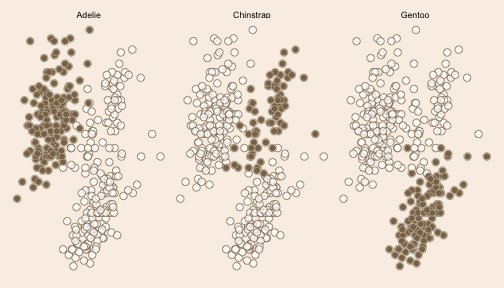<!-- --> ] <style> .panel1-feature-auto { color: black; width: 49%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-feature-auto { color: black; width: 49%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-feature-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- class: center, middle # <span style="color:burlywood">Discussion</span> --- ### In the first layer, *all* data is displayed. This is because we've facetted by species, a variable we've remove via . %>% select(-species) ```r geom_point( * data = . %>% select(-species), size = 3, shape = 21, stroke = .7, color = "burlywood4", fill= "white", alpha = .7 ) ``` --- ### The second layer, shows the category of interest, because the data it uses is the *globally declared data*, which contains the species faceting variable. ### Colors selection draws our attention to this layer. A point with shape = 21, is a circle with a perimiter so a fill color and boarder color may be specified. ```r geom_point( size = 3, shape = 21, stroke = .2, * color = "white", fill = "burlywood4" ) ``` --- <img src="facet-ref-shadow_files/figure-html/unnamed-chunk-3-1.png" width="100%" style="display: block; margin: auto;" /> <style type="text/css"> .remark-code{line-height: 1.5; font-size: 100%} @media print { .has-continuation { display: block; } } </style>