class: center, middle, inverse, title-slide # Exploded code ## Using flipbookr and xaringan ### Me --- <style type="text/css"> .remark-code{line-height: 1.5; font-size: 70%} @media print { .has-continuation { display: block; } } code.r.hljs.remark-code{ position: relative; overflow-x: hidden; } code.r.hljs.remark-code:hover{ overflow-x:visible; width: 500px; border-style: solid; } </style> --- count: false .panel1-setup-auto[ ```r *knitr::opts_chunk$set(echo = TRUE, warning = F, message = F) ``` ] .panel2-setup-auto[ ] <style> .panel1-setup-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-setup-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-setup-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-e1-auto[ ```r *library(tidyverse) ``` ] .panel2-e1-auto[ ] --- count: false .panel1-e1-auto[ ```r library(tidyverse) *library(ggseg) ``` ] .panel2-e1-auto[ ] --- count: false .panel1-e1-auto[ ```r library(tidyverse) library(ggseg) *ggseg( * view = "medial", * ) ``` ] .panel2-e1-auto[ 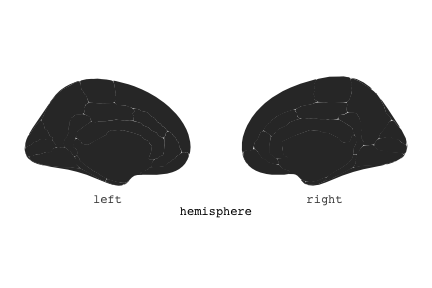<!-- --> ] --- count: false .panel1-e1-auto[ ```r library(tidyverse) library(ggseg) ggseg( view = "medial", ) + * theme_darkbrain() ``` ] .panel2-e1-auto[ 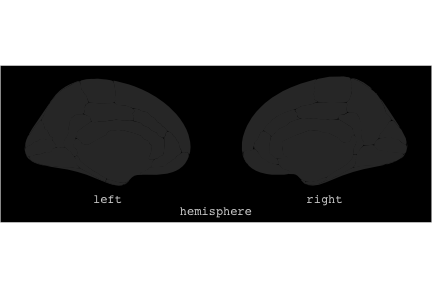<!-- --> ] --- count: false .panel1-e1-auto[ ```r library(tidyverse) library(ggseg) ggseg( view = "medial", ) + theme_darkbrain() + * labs(title = "dk", * subtitle = "medial view") ``` ] .panel2-e1-auto[ 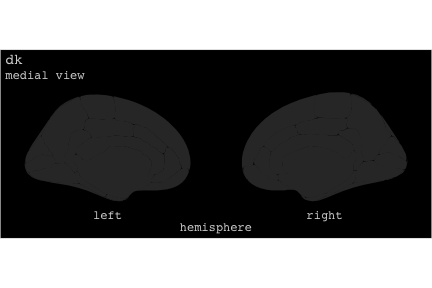<!-- --> ] <style> .panel1-e1-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-e1-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-e1-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-ex2-auto[ ```r *tibble(region = rep(c("transverse temporal", * "insula", * "precentral", * "superior parietal"), 2), * p = sample(seq(0, .5, .001), 8), * g = c("A", "B", "C", "D", "B", "A", "C","A"), * AgeG = c(rep("Young", 4), * rep("Old", 4 )) *) ``` ] .panel2-ex2-auto[ ``` # A tibble: 8 × 4 region p g AgeG <chr> <dbl> <chr> <chr> 1 transverse temporal 0.016 A Young 2 insula 0.215 B Young 3 precentral 0.302 C Young 4 superior parietal 0.276 D Young 5 transverse temporal 0.442 B Old 6 insula 0.323 A Old 7 precentral 0.206 C Old 8 superior parietal 0.322 A Old ``` ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% * ggseg(.data = ., * atlas = dk, * position = "stacked", * show.legend = FALSE) ``` ] .panel2-ex2-auto[ 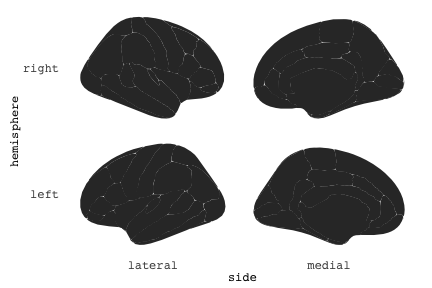<!-- --> ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + * aes(fill = p) ``` ] .panel2-ex2-auto[ 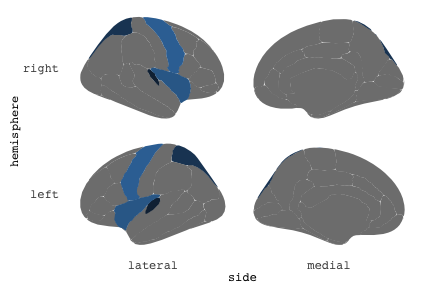<!-- --> ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + * scale_fill_viridis_c() ``` ] .panel2-ex2-auto[ 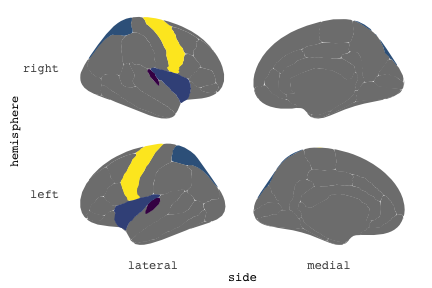<!-- --> ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + * aes(color = g) ``` ] .panel2-ex2-auto[ 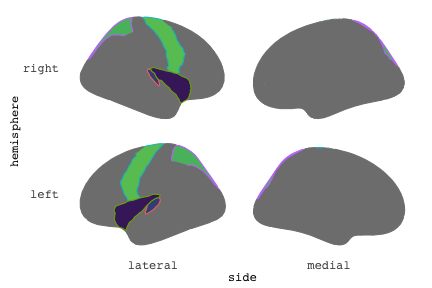<!-- --> ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + aes(color = g) + * scale_colour_viridis_d(option = "magma") ``` ] .panel2-ex2-auto[ 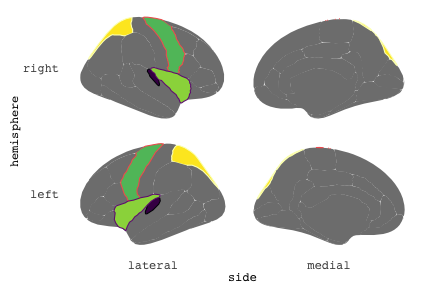<!-- --> ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + aes(color = g) + scale_colour_viridis_d(option = "magma") + * aes(fill = NULL) ``` ] .panel2-ex2-auto[ 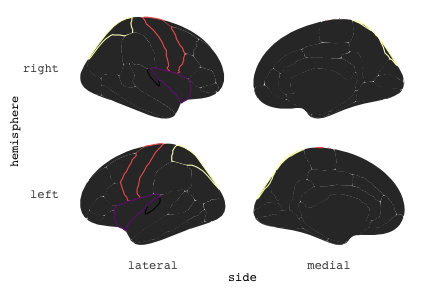<!-- --> ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + aes(color = g) + scale_colour_viridis_d(option = "magma") + aes(fill = NULL) + * facet_wrap(~AgeG, ncol = 3) ``` ] .panel2-ex2-auto[ 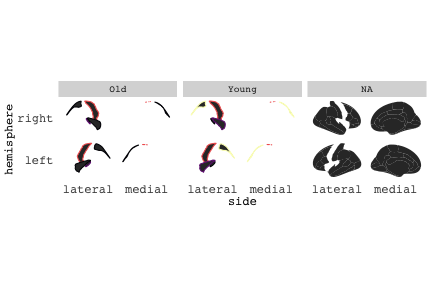<!-- --> ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + aes(color = g) + scale_colour_viridis_d(option = "magma") + aes(fill = NULL) + facet_wrap(~AgeG, ncol = 3) + * labs(title = "Ungrouped data") ``` ] .panel2-ex2-auto[ 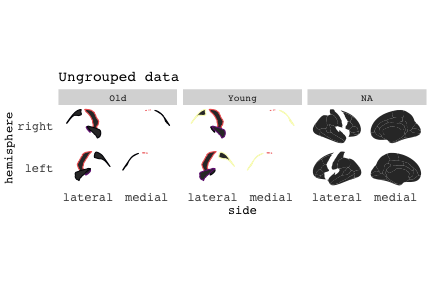<!-- --> ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + aes(color = g) + scale_colour_viridis_d(option = "magma") + aes(fill = NULL) + facet_wrap(~AgeG, ncol = 3) + labs(title = "Ungrouped data") *layer_data(plot = last_plot()) ``` ] .panel2-ex2-auto[ 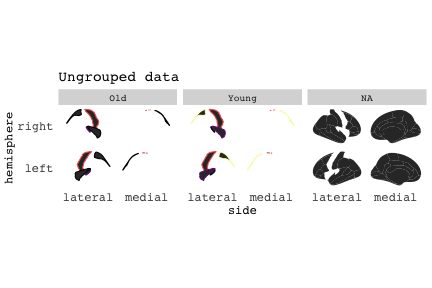<!-- --> ``` colour y x group subgroup PANEL fill size linetype alpha 1 <NA> 34.46407 84.32563 1 1 3 grey20 0.5 1 NA 2 <NA> 33.33467 84.16625 1 1 3 grey20 0.5 1 NA 3 <NA> 26.38733 92.88468 1 1 3 grey20 0.5 1 NA 4 <NA> 27.83508 92.55951 1 1 3 grey20 0.5 1 NA 5 <NA> 32.99130 88.42844 1 1 3 grey20 0.5 1 NA 6 <NA> 34.31282 86.28085 1 1 3 grey20 0.5 1 NA 7 <NA> 34.46407 84.32563 1 1 3 grey20 0.5 1 NA 8 <NA> 23.91232 94.20152 2 1 3 grey20 0.5 1 NA 9 <NA> 23.90330 94.20817 2 1 3 grey20 0.5 1 NA 10 <NA> 23.91323 94.21681 2 1 3 grey20 0.5 1 NA 11 <NA> 23.92040 94.21003 2 1 3 grey20 0.5 1 NA 12 <NA> 23.91232 94.20152 2 1 3 grey20 0.5 1 NA 13 <NA> 108.81394 214.82147 3 1 3 grey20 0.5 1 NA 14 <NA> 107.65361 210.46947 3 1 3 grey20 0.5 1 NA 15 <NA> 104.40110 207.95788 3 1 3 grey20 0.5 1 NA 16 <NA> 101.39895 204.07948 3 1 3 grey20 0.5 1 NA 17 <NA> 89.95142 195.91792 3 1 3 grey20 0.5 1 NA 18 <NA> 86.66585 189.97995 3 1 3 grey20 0.5 1 NA 19 <NA> 83.26921 186.62237 3 1 3 grey20 0.5 1 NA 20 <NA> 73.29106 180.72329 3 1 3 grey20 0.5 1 NA 21 <NA> 65.48774 180.03683 3 1 3 grey20 0.5 1 NA 22 <NA> 61.54807 177.14671 3 1 3 grey20 0.5 1 NA 23 <NA> 60.99465 181.17933 3 1 3 grey20 0.5 1 NA 24 <NA> 61.81593 191.39561 3 1 3 grey20 0.5 1 NA 25 <NA> 64.18767 206.04892 3 1 3 grey20 0.5 1 NA 26 <NA> 65.36395 209.24998 3 1 3 grey20 0.5 1 NA 27 <NA> 70.32102 217.20044 3 1 3 grey20 0.5 1 NA 28 <NA> 73.19604 220.21282 3 1 3 grey20 0.5 1 NA 29 <NA> 88.53093 230.62533 3 1 3 grey20 0.5 1 NA 30 <NA> 90.22672 230.99573 3 1 3 grey20 0.5 1 NA 31 <NA> 98.13730 227.17123 3 1 3 grey20 0.5 1 NA 32 <NA> 101.07891 222.87775 3 1 3 grey20 0.5 1 NA 33 <NA> 103.63107 220.61792 3 1 3 grey20 0.5 1 NA 34 <NA> 104.69531 217.45709 3 1 3 grey20 0.5 1 NA 35 <NA> 108.81394 214.82147 3 1 3 grey20 0.5 1 NA 36 <NA> 184.31442 106.16003 4 1 3 grey20 0.5 1 NA 37 <NA> 180.91132 93.61281 4 1 3 grey20 0.5 1 NA 38 <NA> 173.94538 76.94933 4 1 3 grey20 0.5 1 NA 39 <NA> 168.89276 67.47581 4 1 3 grey20 0.5 1 NA 40 <NA> 163.11316 58.86546 4 1 3 grey20 0.5 1 NA 41 <NA> 157.50335 52.76675 4 1 3 grey20 0.5 1 NA 42 <NA> 156.46844 52.69114 4 1 3 grey20 0.5 1 NA 43 <NA> 154.55683 54.85056 4 1 3 grey20 0.5 1 NA 44 <NA> 151.74032 55.37262 4 1 3 grey20 0.5 1 NA 45 <NA> 149.33529 57.62704 4 1 3 grey20 0.5 1 NA 46 <NA> 144.95095 57.35486 4 1 3 grey20 0.5 1 NA 47 <NA> 141.50689 60.59761 4 1 3 grey20 0.5 1 NA 48 <NA> 136.85295 61.11059 4 1 3 grey20 0.5 1 NA 49 <NA> 132.21793 63.30225 4 1 3 grey20 0.5 1 NA 50 <NA> 128.95512 66.98750 4 1 3 grey20 0.5 1 NA 51 <NA> 124.05270 69.23790 4 1 3 grey20 0.5 1 NA 52 <NA> 123.23892 73.82631 4 1 3 grey20 0.5 1 NA 53 <NA> 126.11653 80.95370 4 1 3 grey20 0.5 1 NA 54 <NA> 133.15302 85.83042 4 1 3 grey20 0.5 1 NA 55 <NA> 141.51071 88.70268 4 1 3 grey20 0.5 1 NA 56 <NA> 146.76656 93.26094 4 1 3 grey20 0.5 1 NA 57 <NA> 157.19585 97.90184 4 1 3 grey20 0.5 1 NA 58 <NA> 166.38766 103.49982 4 1 3 grey20 0.5 1 NA 59 <NA> 170.97680 104.68918 4 1 3 grey20 0.5 1 NA 60 <NA> 178.06644 111.59143 4 1 3 grey20 0.5 1 NA 61 <NA> 180.56341 113.08734 4 1 3 grey20 0.5 1 NA 62 <NA> 182.70929 113.52920 4 1 3 grey20 0.5 1 NA 63 <NA> 183.95871 112.36570 4 1 3 grey20 0.5 1 NA 64 <NA> 184.31442 106.16003 4 1 3 grey20 0.5 1 NA 65 <NA> 48.35713 256.54807 5 1 3 grey20 0.5 1 NA 66 <NA> 45.02750 244.04403 5 1 3 grey20 0.5 1 NA 67 <NA> 33.05511 221.35735 5 1 3 grey20 0.5 1 NA 68 <NA> 33.11799 224.62905 5 1 3 grey20 0.5 1 NA 69 <NA> 41.05674 249.18920 5 1 3 grey20 0.5 1 NA 70 <NA> 45.47859 262.31775 5 1 3 grey20 0.5 1 NA 71 <NA> 47.16155 264.32461 5 1 3 grey20 0.5 1 NA 72 <NA> 48.33476 263.21036 5 1 3 grey20 0.5 1 NA 73 <NA> 48.35713 256.54807 5 1 3 grey20 0.5 1 NA 74 <NA> 31.75113 219.14196 6 1 3 grey20 0.5 1 NA 75 <NA> 31.74434 219.15742 6 1 3 grey20 0.5 1 NA 76 <NA> 31.75275 219.17732 6 1 3 grey20 0.5 1 NA 77 <NA> 31.75961 219.16237 6 1 3 grey20 0.5 1 NA 78 <NA> 31.75113 219.14196 6 1 3 grey20 0.5 1 NA 79 <NA> 30.77113 217.18196 7 1 3 grey20 0.5 1 NA 80 <NA> 30.76434 217.19742 7 1 3 grey20 0.5 1 NA 81 <NA> 30.77275 217.21732 7 1 3 grey20 0.5 1 NA 82 <NA> 30.77961 217.20237 7 1 3 grey20 0.5 1 NA 83 <NA> 30.77113 217.18196 7 1 3 grey20 0.5 1 NA 84 <NA> 161.62334 218.43732 8 1 3 grey20 0.5 1 NA 85 <NA> 157.68995 214.91403 8 1 3 grey20 0.5 1 NA 86 <NA> 152.43575 223.29315 8 1 3 grey20 0.5 1 NA 87 <NA> 149.66529 225.30004 8 1 3 grey20 0.5 1 NA 88 <NA> 138.37893 224.17000 8 1 3 grey20 0.5 1 NA 89 <NA> 126.96534 225.61354 8 1 3 grey20 0.5 1 NA 90 <NA> 117.54399 223.65238 8 1 3 grey20 0.5 1 NA 91 <NA> 112.99908 218.37809 8 1 3 grey20 0.5 1 NA 92 <NA> 109.54029 216.32304 8 1 3 grey20 0.5 1 NA 93 <NA> 106.89191 216.03190 8 1 3 grey20 0.5 1 NA 94 <NA> 104.23573 217.24576 8 1 3 grey20 0.5 1 NA 95 <NA> 98.48910 226.26403 8 1 3 grey20 0.5 1 NA 96 <NA> 90.26738 231.86209 8 1 3 grey20 0.5 1 NA 97 <NA> 89.88409 238.72863 8 1 3 grey20 0.5 1 NA 98 <NA> 87.17573 241.77224 8 1 3 grey20 0.5 1 NA 99 <NA> 83.16538 249.06756 8 1 3 grey20 0.5 1 NA 100 <NA> 84.48085 252.91240 8 1 3 grey20 0.5 1 NA [ reached 'max' / getOption("max.print") -- omitted 3137 rows ] ``` ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + aes(color = g) + scale_colour_viridis_d(option = "magma") + aes(fill = NULL) + facet_wrap(~AgeG, ncol = 3) + labs(title = "Ungrouped data") layer_data(plot = last_plot()) -> * hide ``` ] .panel2-ex2-auto[ 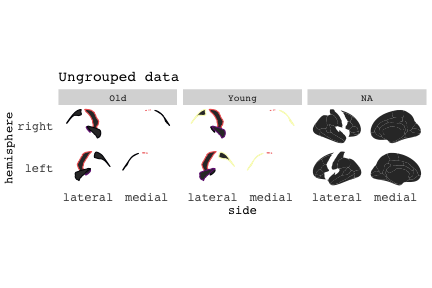<!-- --> ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + aes(color = g) + scale_colour_viridis_d(option = "magma") + aes(fill = NULL) + facet_wrap(~AgeG, ncol = 3) + labs(title = "Ungrouped data") layer_data(plot = last_plot()) -> hide *ggseg ``` ] .panel2-ex2-auto[ 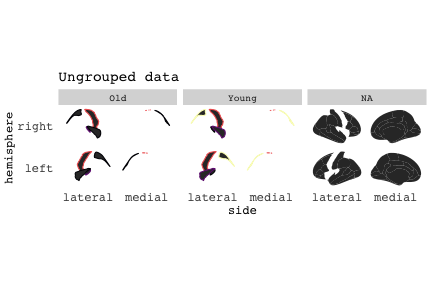<!-- --> ``` function (.data = NULL, atlas = "dk", position = "dispersed", view = NULL, hemisphere = NULL, adapt_scales = TRUE, ...) { atlas <- if (!is.character(atlas)) { atlas } else { get(atlas) } if (!is_ggseg_atlas(atlas)) atlas <- as_ggseg_atlas(atlas) atlas <- unnest(atlas, ggseg) stack <- match.arg(position, c("stacked", "dispersed"), several.ok = FALSE) if (stack == "stacked") { atlas <- stack_brain(atlas) } if (!is.null(hemisphere)) atlas <- filter(atlas, hemi %in% hemisphere) if (!is.null(view)) { atlas <- filter(atlas, grepl(view, side)) if (view == "lateral" & (all(c("left", "right") %in% hemisphere) | is.null(hemisphere)) & stack == "dispersed") { atlas <- squish_position(atlas, hemisphere) } } if (!is.null(.data)) { if (is_brain_atlas(.data) | is_ggseg_atlas(.data)) stop("Atlas given as '.data', did you mean to give it to 'atlas'?") atlas <- brain_join(.data, atlas) atlas <- filter(atlas, !is.na(.long)) } gg <- ggplot(data = atlas, aes(x = .long, y = .lat, group = .id, subgroup = .subid)) + geom_polygon(...) + coord_fixed() if (adapt_scales) { gg <- gg + scale_y_brain(atlas, stack) + scale_x_brain(atlas, stack) + scale_labs_brain(atlas, stack) } gg + theme_brain() } <bytecode: 0x7fef8c773b90> <environment: namespace:ggseg> ``` ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + aes(color = g) + scale_colour_viridis_d(option = "magma") + aes(fill = NULL) + facet_wrap(~AgeG, ncol = 3) + labs(title = "Ungrouped data") layer_data(plot = last_plot()) -> hide ggseg -> * hide ``` ] .panel2-ex2-auto[ 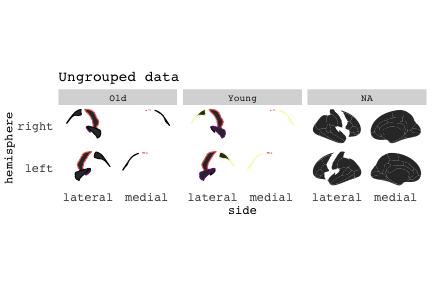<!-- --> ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + aes(color = g) + scale_colour_viridis_d(option = "magma") + aes(fill = NULL) + facet_wrap(~AgeG, ncol = 3) + labs(title = "Ungrouped data") layer_data(plot = last_plot()) -> hide ggseg -> hide *dk ``` ] .panel2-ex2-auto[ 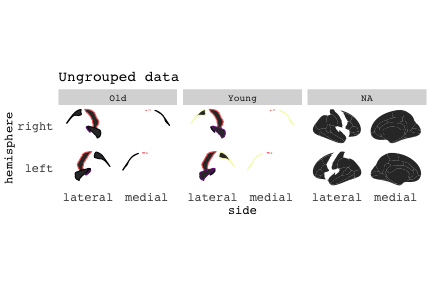<!-- --> ``` # dk cortical brain atlas regions: 35 hemispheres: left, right side views: lateral, medial palette: yes use: ggplot() + geom_brain() ---- hemi side region label roi <chr> <chr> <chr> <chr> <chr> 1 left lateral bankssts lh_bankssts 0002 2 left lateral caudal middle frontal lh_caudalmiddlefrontal 0004 3 left lateral fusiform lh_fusiform 0008 4 left lateral inferior parietal lh_inferiorparietal 0009 5 left lateral inferior temporal lh_inferiortemporal 0010 6 left lateral lateral occipital lh_lateraloccipital 0012 7 left lateral lateral orbitofrontal lh_lateralorbitofrontal 0013 8 left lateral middle temporal lh_middletemporal 0016 9 left lateral pars opercularis lh_parsopercularis 0019 10 left lateral pars orbitalis lh_parsorbitalis 0020 11 left lateral pars triangularis lh_parstriangularis 0021 12 left lateral postcentral lh_postcentral 0023 13 left lateral precentral lh_precentral 0025 14 left lateral rostral middle frontal lh_rostralmiddlefrontal 0028 15 left lateral superior frontal lh_superiorfrontal 0029 16 left lateral superior parietal lh_superiorparietal 0030 17 left lateral superior temporal lh_superiortemporal 0031 18 left lateral supramarginal lh_supramarginal 0032 19 left lateral temporal pole lh_temporalpole 0034 20 left lateral transverse temporal lh_transversetemporal 0035 21 left lateral insula lh_insula 0036 22 left medial caudal anterior cingulate lh_caudalanteriorcingulate 0003 23 left medial corpus callosum lh_corpuscallosum 0005 24 left medial cuneus lh_cuneus 0006 25 left medial entorhinal lh_entorhinal 0007 26 left medial fusiform lh_fusiform 0008 27 left medial isthmus cingulate lh_isthmuscingulate 0011 28 left medial lateral occipital lh_lateraloccipital 0012 29 left medial lateral orbitofrontal lh_lateralorbitofrontal 0013 30 left medial lingual lh_lingual 0014 31 left medial medial orbitofrontal lh_medialorbitofrontal 0015 32 left medial parahippocampal lh_parahippocampal 0017 33 left medial paracentral lh_paracentral 0018 34 left medial pericalcarine lh_pericalcarine 0022 35 left medial postcentral lh_postcentral 0023 36 left medial posterior cingulate lh_posteriorcingulate 0024 37 left medial precentral lh_precentral 0025 38 left medial precuneus lh_precuneus 0026 39 left medial rostral anterior cingulate lh_rostralanteriorcingulate 0027 40 left medial superior frontal lh_superiorfrontal 0029 41 left medial superior parietal lh_superiorparietal 0030 42 left medial frontal pole lh_frontalpole 0033 43 left medial temporal pole lh_temporalpole 0034 44 right medial caudal anterior cingulate rh_caudalanteriorcingulate 0003 45 right medial corpus callosum rh_corpuscallosum 0005 46 right medial cuneus rh_cuneus 0006 47 right medial entorhinal rh_entorhinal 0007 48 right medial fusiform rh_fusiform 0008 49 right medial isthmus cingulate rh_isthmuscingulate 0011 50 right medial lateral occipital rh_lateraloccipital 0012 51 right medial lateral orbitofrontal rh_lateralorbitofrontal 0013 52 right medial lingual rh_lingual 0014 53 right medial medial orbitofrontal rh_medialorbitofrontal 0015 54 right medial parahippocampal rh_parahippocampal 0017 55 right medial paracentral rh_paracentral 0018 # … with 31 more rows ``` ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + aes(color = g) + scale_colour_viridis_d(option = "magma") + aes(fill = NULL) + facet_wrap(~AgeG, ncol = 3) + labs(title = "Ungrouped data") layer_data(plot = last_plot()) -> hide ggseg -> hide dk -> * hide ``` ] .panel2-ex2-auto[ 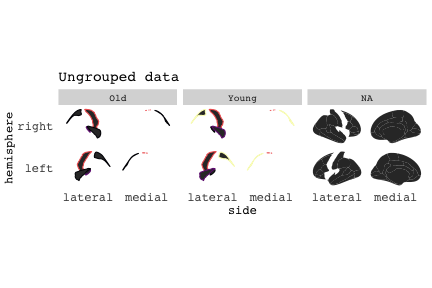<!-- --> ] --- count: false .panel1-ex2-auto[ ```r tibble(region = rep(c("transverse temporal", "insula", "precentral", "superior parietal"), 2), p = sample(seq(0, .5, .001), 8), g = c("A", "B", "C", "D", "B", "A", "C","A"), AgeG = c(rep("Young", 4), rep("Old", 4 )) ) %>% ggseg(.data = ., atlas = dk, position = "stacked", show.legend = FALSE) + aes(fill = p) + scale_fill_viridis_c() + aes(color = g) + scale_colour_viridis_d(option = "magma") + aes(fill = NULL) + facet_wrap(~AgeG, ncol = 3) + labs(title = "Ungrouped data") layer_data(plot = last_plot()) -> hide ggseg -> hide dk -> hide ``` ] .panel2-ex2-auto[ 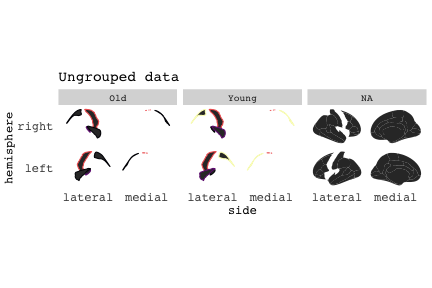<!-- --> ] <style> .panel1-ex2-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-ex2-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-ex2-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-ex3-auto[ ```r *ggseg (atlas = aseg) ``` ] .panel2-ex3-auto[ 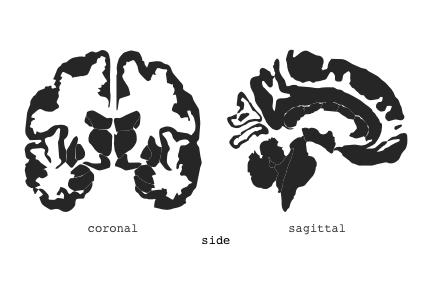<!-- --> ] --- count: false .panel1-ex3-auto[ ```r ggseg (atlas = aseg) + * aes(fill = region) ``` ] .panel2-ex3-auto[ 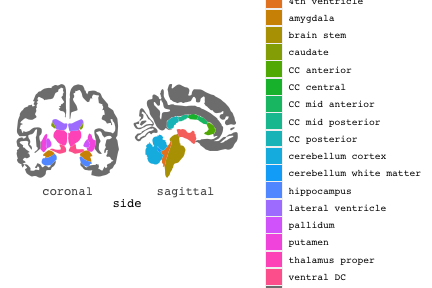<!-- --> ] --- count: false .panel1-ex3-auto[ ```r ggseg (atlas = aseg) + aes(fill = region) + * theme(legend.justification = c(1, 0)) ``` ] .panel2-ex3-auto[ <!-- --> ] --- count: false .panel1-ex3-auto[ ```r ggseg (atlas = aseg) + aes(fill = region) + theme(legend.justification = c(1, 0)) + * theme(legend.position = "bottom") ``` ] .panel2-ex3-auto[ <!-- --> ] --- count: false .panel1-ex3-auto[ ```r ggseg (atlas = aseg) + aes(fill = region) + theme(legend.justification = c(1, 0)) + theme(legend.position = "bottom") + * theme(legend.text = element_text(size = 5)) ``` ] .panel2-ex3-auto[ <!-- --> ] --- count: false .panel1-ex3-auto[ ```r ggseg (atlas = aseg) + aes(fill = region) + theme(legend.justification = c(1, 0)) + theme(legend.position = "bottom") + theme(legend.text = element_text(size = 5)) + * guides(fill = guide_legend(ncol = 3)) ``` ] .panel2-ex3-auto[ <!-- --> ] --- count: false .panel1-ex3-auto[ ```r ggseg (atlas = aseg) + aes(fill = region) + theme(legend.justification = c(1, 0)) + theme(legend.position = "bottom") + theme(legend.text = element_text(size = 5)) + guides(fill = guide_legend(ncol = 3)) *layer_data(plot = last_plot()) ``` ] .panel2-ex3-auto[ 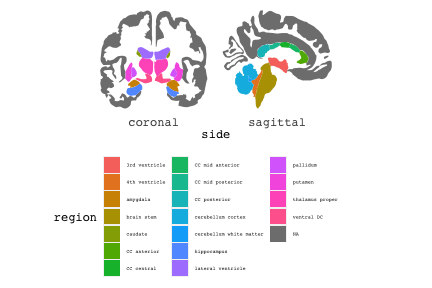<!-- --> ``` fill y x group subgroup PANEL colour size linetype alpha 1 grey50 2.70275 2.07673 1 1 1 NA 0.5 1 NA 2 grey50 2.70389 2.07683 1 1 1 NA 0.5 1 NA 3 grey50 2.70801 2.07043 1 1 1 NA 0.5 1 NA 4 grey50 2.71468 2.05593 1 1 1 NA 0.5 1 NA 5 grey50 2.72005 2.04113 1 1 1 NA 0.5 1 NA 6 grey50 2.72201 2.03423 1 1 1 NA 0.5 1 NA 7 grey50 2.72152 2.03393 1 1 1 NA 0.5 1 NA 8 grey50 2.71507 2.03013 1 1 1 NA 0.5 1 NA 9 grey50 2.64708 1.99823 1 1 1 NA 0.5 1 NA 10 grey50 2.64008 1.99553 1 1 1 NA 0.5 1 NA 11 grey50 2.63748 1.99453 1 1 1 NA 0.5 1 NA 12 grey50 2.62050 1.99383 1 1 1 NA 0.5 1 NA 13 grey50 2.60743 1.99253 1 1 1 NA 0.5 1 NA 14 grey50 2.60672 1.99123 1 1 1 NA 0.5 1 NA 15 grey50 2.60461 1.98683 1 1 1 NA 0.5 1 NA 16 grey50 2.59809 1.97363 1 1 1 NA 0.5 1 NA 17 grey50 2.58979 1.95483 1 1 1 NA 0.5 1 NA 18 grey50 2.58497 1.93763 1 1 1 NA 0.5 1 NA 19 grey50 2.58518 1.92713 1 1 1 NA 0.5 1 NA 20 grey50 2.58675 1.92473 1 1 1 NA 0.5 1 NA 21 grey50 2.59240 1.91583 1 1 1 NA 0.5 1 NA 22 grey50 2.62696 1.87533 1 1 1 NA 0.5 1 NA 23 grey50 2.66597 1.83793 1 1 1 NA 0.5 1 NA 24 grey50 2.67372 1.83053 1 1 1 NA 0.5 1 NA 25 grey50 2.68077 1.82393 1 1 1 NA 0.5 1 NA 26 grey50 2.71632 1.79013 1 1 1 NA 0.5 1 NA 27 grey50 2.75477 1.76153 1 1 1 NA 0.5 1 NA 28 grey50 2.76325 1.75753 1 1 1 NA 0.5 1 NA 29 grey50 2.77225 1.75323 1 1 1 NA 0.5 1 NA 30 grey50 2.82205 1.74173 1 1 1 NA 0.5 1 NA 31 grey50 2.87257 1.73143 1 1 1 NA 0.5 1 NA 32 grey50 2.88189 1.72753 1 1 1 NA 0.5 1 NA 33 grey50 2.89226 1.72333 1 1 1 NA 0.5 1 NA 34 grey50 2.94390 1.69833 1 1 1 NA 0.5 1 NA 35 grey50 2.98909 1.66553 1 1 1 NA 0.5 1 NA 36 grey50 2.99626 1.65763 1 1 1 NA 0.5 1 NA 37 grey50 2.99837 1.65523 1 1 1 NA 0.5 1 NA 38 grey50 2.99811 1.64243 1 1 1 NA 0.5 1 NA 39 grey50 2.99045 1.61893 1 1 1 NA 0.5 1 NA 40 grey50 2.97873 1.59183 1 1 1 NA 0.5 1 NA 41 grey50 2.97043 1.57253 1 1 1 NA 0.5 1 NA 42 grey50 2.96815 1.56593 1 1 1 NA 0.5 1 NA 43 grey50 2.96761 1.56423 1 1 1 NA 0.5 1 NA 44 grey50 2.96180 1.55543 1 1 1 NA 0.5 1 NA 45 grey50 2.95807 1.54813 1 1 1 NA 0.5 1 NA 46 grey50 2.95850 1.54723 1 1 1 NA 0.5 1 NA 47 grey50 2.96111 1.54213 1 1 1 NA 0.5 1 NA 48 grey50 2.96848 1.52653 1 1 1 NA 0.5 1 NA 49 grey50 2.96925 1.51603 1 1 1 NA 0.5 1 NA 50 grey50 2.95833 1.51153 1 1 1 NA 0.5 1 NA 51 grey50 2.93957 1.50963 1 1 1 NA 0.5 1 NA 52 grey50 2.93327 1.50913 1 1 1 NA 0.5 1 NA 53 grey50 2.93007 1.50893 1 1 1 NA 0.5 1 NA 54 grey50 2.91157 1.50213 1 1 1 NA 0.5 1 NA 55 grey50 2.89827 1.49113 1 1 1 NA 0.5 1 NA 56 grey50 2.89768 1.48883 1 1 1 NA 0.5 1 NA 57 grey50 2.89594 1.48193 1 1 1 NA 0.5 1 NA 58 grey50 2.88911 1.46163 1 1 1 NA 0.5 1 NA 59 grey50 2.87727 1.44653 1 1 1 NA 0.5 1 NA 60 grey50 2.85868 1.44563 1 1 1 NA 0.5 1 NA 61 grey50 2.83632 1.45643 1 1 1 NA 0.5 1 NA 62 grey50 2.82954 1.46113 1 1 1 NA 0.5 1 NA 63 grey50 2.82487 1.46433 1 1 1 NA 0.5 1 NA 64 grey50 2.80757 1.46913 1 1 1 NA 0.5 1 NA 65 grey50 2.77936 1.47103 1 1 1 NA 0.5 1 NA 66 grey50 2.75261 1.46683 1 1 1 NA 0.5 1 NA 67 grey50 2.73779 1.46043 1 1 1 NA 0.5 1 NA 68 grey50 2.73470 1.45673 1 1 1 NA 0.5 1 NA 69 grey50 2.72884 1.44973 1 1 1 NA 0.5 1 NA 70 grey50 2.70514 1.43373 1 1 1 NA 0.5 1 NA 71 grey50 2.67622 1.43013 1 1 1 NA 0.5 1 NA 72 grey50 2.64887 1.43803 1 1 1 NA 0.5 1 NA 73 grey50 2.63021 1.44683 1 1 1 NA 0.5 1 NA 74 grey50 2.62397 1.44973 1 1 1 NA 0.5 1 NA 75 grey50 2.60991 1.45623 1 1 1 NA 0.5 1 NA 76 grey50 2.54390 1.49833 1 1 1 NA 0.5 1 NA 77 grey50 2.47797 1.54063 1 1 1 NA 0.5 1 NA 78 grey50 2.46393 1.54743 1 1 1 NA 0.5 1 NA 79 grey50 2.45269 1.55273 1 1 1 NA 0.5 1 NA 80 grey50 2.39226 1.57113 1 1 1 NA 0.5 1 NA 81 grey50 2.32948 1.58373 1 1 1 NA 0.5 1 NA 82 grey50 2.31701 1.58633 1 1 1 NA 0.5 1 NA 83 grey50 2.31576 1.58653 1 1 1 NA 0.5 1 NA 84 grey50 2.30566 1.57403 1 1 1 NA 0.5 1 NA 85 grey50 2.29573 1.55443 1 1 1 NA 0.5 1 NA 86 grey50 2.29459 1.55113 1 1 1 NA 0.5 1 NA 87 grey50 2.28987 1.53713 1 1 1 NA 0.5 1 NA 88 grey50 2.26775 1.46643 1 1 1 NA 0.5 1 NA 89 grey50 2.24415 1.39653 1 1 1 NA 0.5 1 NA 90 grey50 2.23861 1.38283 1 1 1 NA 0.5 1 NA 91 grey50 2.23747 1.37983 1 1 1 NA 0.5 1 NA 92 grey50 2.21875 1.36933 1 1 1 NA 0.5 1 NA 93 grey50 2.19352 1.36303 1 1 1 NA 0.5 1 NA 94 grey50 2.18908 1.36223 1 1 1 NA 0.5 1 NA 95 grey50 2.17557 1.35983 1 1 1 NA 0.5 1 NA 96 grey50 2.03983 1.33963 1 1 1 NA 0.5 1 NA 97 grey50 2.02626 1.33743 1 1 1 NA 0.5 1 NA 98 grey50 2.02420 1.33703 1 1 1 NA 0.5 1 NA 99 grey50 2.01465 1.33053 1 1 1 NA 0.5 1 NA 100 grey50 2.00548 1.32183 1 1 1 NA 0.5 1 NA [ reached 'max' / getOption("max.print") -- omitted 5085 rows ] ``` ] --- count: false .panel1-ex3-auto[ ```r ggseg (atlas = aseg) + aes(fill = region) + theme(legend.justification = c(1, 0)) + theme(legend.position = "bottom") + theme(legend.text = element_text(size = 5)) + guides(fill = guide_legend(ncol = 3)) layer_data(plot = last_plot()) -> * hide ``` ] .panel2-ex3-auto[ 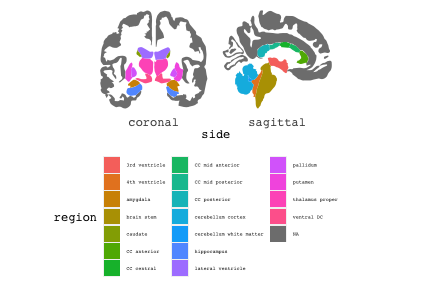<!-- --> ] --- count: false .panel1-ex3-auto[ ```r ggseg (atlas = aseg) + aes(fill = region) + theme(legend.justification = c(1, 0)) + theme(legend.position = "bottom") + theme(legend.text = element_text(size = 5)) + guides(fill = guide_legend(ncol = 3)) layer_data(plot = last_plot()) -> hide *aseg ``` ] .panel2-ex3-auto[ 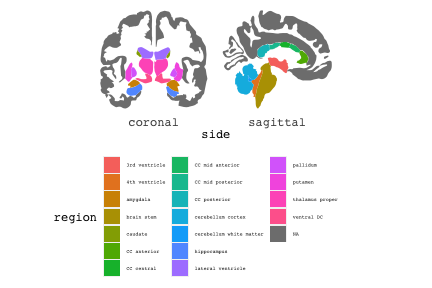<!-- --> ``` # aseg subcortical brain atlas regions: 18 hemispheres: right, left, midline side views: coronal, sagittal palette: yes use: ggplot() + geom_brain() ---- hemi region side label <chr> <chr> <chr> <chr> 1 left thalamus proper coronal Left-Thalamus-Proper 2 right thalamus proper coronal Right-Thalamus-Proper 3 right lateral ventricle coronal Right-Lateral-Ventricle 4 left hippocampus coronal Left-Hippocampus 5 left lateral ventricle coronal Left-Lateral-Ventricle 6 right putamen coronal Right-Putamen 7 right amygdala coronal Right-Amygdala 8 left putamen coronal Left-Putamen 9 right ventral DC coronal Right-VentralDC 10 left ventral DC coronal Left-VentralDC 11 right hippocampus coronal Right-Hippocampus 12 left amygdala coronal Left-Amygdala 13 left pallidum coronal Left-Pallidum 14 right pallidum coronal Right-Pallidum 15 left caudate coronal Left-Caudate 16 right caudate coronal Right-Caudate 17 midline 3rd ventricle sagittal x3rd-ventricle 18 midline 4th ventricle sagittal x4th-ventricle 19 midline brain stem sagittal Brain-Stem 20 midline CC anterior sagittal CC_Anterior 21 midline CC central sagittal CC_Central 22 midline CC mid anterior sagittal CC_Mid_Anterior 23 midline CC mid posterior sagittal CC_Mid_Posterior 24 midline CC posterior sagittal CC_Posterior 25 midline cerebellum cortex sagittal Right-Cerebellum-Cortex 26 midline cerebellum white matter sagittal Right-Cerebellum-White-Matter ``` ] <style> .panel1-ex3-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-ex3-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-ex3-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style>