class: center, middle, inverse, title-slide # ggsample and the CLT ## Using flipbookr and xaringan ### Me --- <style type="text/css"> .remark-code{line-height: 1.5; font-size: 70%} @media print { .has-continuation { display: block; } } code.r.hljs.remark-code{ position: relative; overflow-x: hidden; } code.r.hljs.remark-code:hover{ overflow-x:visible; width: 500px; border-style: solid; } </style> --- --- count: false .panel1-faithful1-auto[ ```r # population mean *ggplot(data = faithful) ``` ] .panel2-faithful1-auto[ <!-- --> ] --- count: false .panel1-faithful1-auto[ ```r # population mean ggplot(data = faithful) + * aes(x = waiting) ``` ] .panel2-faithful1-auto[ <!-- --> ] --- count: false .panel1-faithful1-auto[ ```r # population mean ggplot(data = faithful) + aes(x = waiting) + * geom_rug() ``` ] .panel2-faithful1-auto[ <!-- --> ] --- count: false .panel1-faithful1-auto[ ```r # population mean ggplot(data = faithful) + aes(x = waiting) + geom_rug() + * geom_dotplot(dotsize = .5) ``` ] .panel2-faithful1-auto[ 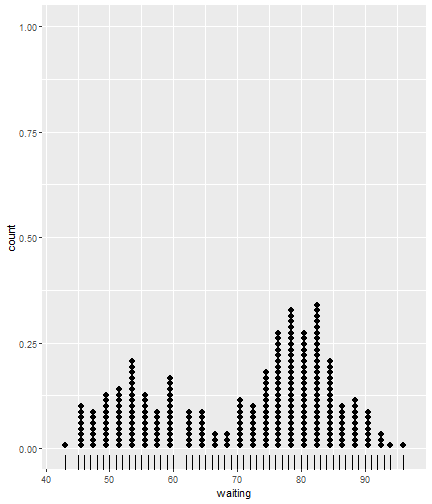<!-- --> ] --- count: false .panel1-faithful1-auto[ ```r # population mean ggplot(data = faithful) + aes(x = waiting) + geom_rug() + geom_dotplot(dotsize = .5) + * ggxmean::geom_x_mean() ``` ] .panel2-faithful1-auto[ 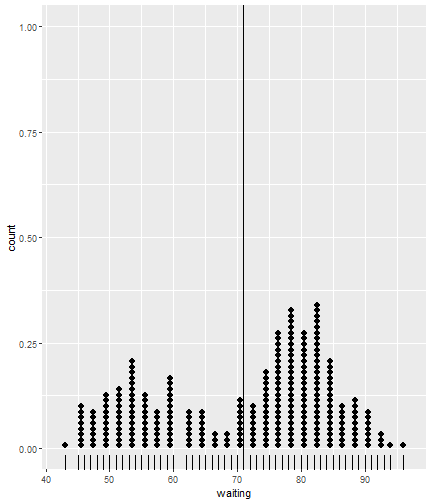<!-- --> ] --- count: false .panel1-faithful1-auto[ ```r # population mean ggplot(data = faithful) + aes(x = waiting) + geom_rug() + geom_dotplot(dotsize = .5) + ggxmean::geom_x_mean() + * ggxmean::geom_x_mean_label() ``` ] .panel2-faithful1-auto[ 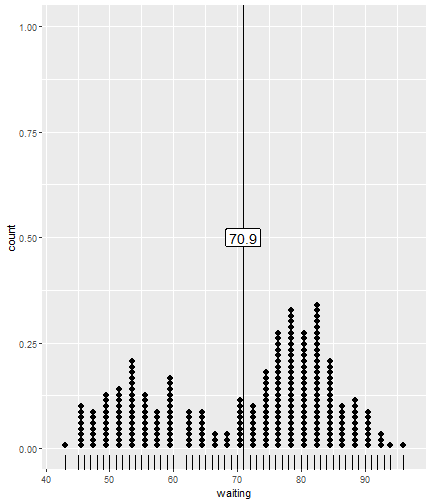<!-- --> ] <style> .panel1-faithful1-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-faithful1-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-faithful1-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-single_sample-10[ ```r # A single sample mean last_plot() + scale_x_continuous(limits = c(40,100)) + ggsample::facet_sample(n_facets = 1, n_sampled = 20) ``` ] .panel2-single_sample-10[ 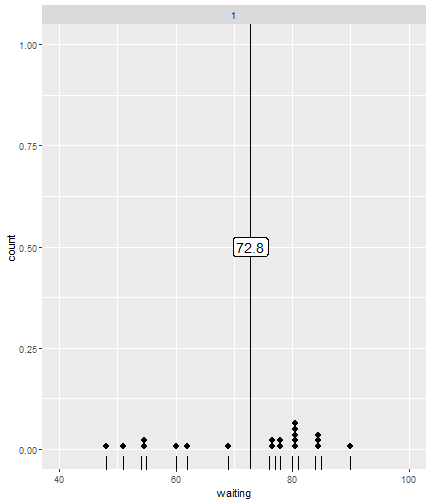<!-- --> ] --- count: false .panel1-single_sample-10[ ```r # A single sample mean last_plot() + scale_x_continuous(limits = c(40,100)) + ggsample::facet_sample(n_facets = 1, n_sampled = 20) ``` ] .panel2-single_sample-10[ <!-- --> ] --- count: false .panel1-single_sample-10[ ```r # A single sample mean last_plot() + scale_x_continuous(limits = c(40,100)) + ggsample::facet_sample(n_facets = 1, n_sampled = 20) ``` ] .panel2-single_sample-10[ <!-- --> ] --- count: false .panel1-single_sample-10[ ```r # A single sample mean last_plot() + scale_x_continuous(limits = c(40,100)) + ggsample::facet_sample(n_facets = 1, n_sampled = 20) ``` ] .panel2-single_sample-10[ <!-- --> ] --- count: false .panel1-single_sample-10[ ```r # A single sample mean last_plot() + scale_x_continuous(limits = c(40,100)) + ggsample::facet_sample(n_facets = 1, n_sampled = 20) ``` ] .panel2-single_sample-10[ <!-- --> ] --- count: false .panel1-single_sample-10[ ```r # A single sample mean last_plot() + scale_x_continuous(limits = c(40,100)) + ggsample::facet_sample(n_facets = 1, n_sampled = 20) ``` ] .panel2-single_sample-10[ <!-- --> ] --- count: false .panel1-single_sample-10[ ```r # A single sample mean last_plot() + scale_x_continuous(limits = c(40,100)) + ggsample::facet_sample(n_facets = 1, n_sampled = 20) ``` ] .panel2-single_sample-10[ <!-- --> ] --- count: false .panel1-single_sample-10[ ```r # A single sample mean last_plot() + scale_x_continuous(limits = c(40,100)) + ggsample::facet_sample(n_facets = 1, n_sampled = 20) ``` ] .panel2-single_sample-10[ <!-- --> ] --- count: false .panel1-single_sample-10[ ```r # A single sample mean last_plot() + scale_x_continuous(limits = c(40,100)) + ggsample::facet_sample(n_facets = 1, n_sampled = 20) ``` ] .panel2-single_sample-10[ <!-- --> ] --- count: false .panel1-single_sample-10[ ```r # A single sample mean last_plot() + scale_x_continuous(limits = c(40,100)) + ggsample::facet_sample(n_facets = 1, n_sampled = 20) ``` ] .panel2-single_sample-10[ <!-- --> ] <style> .panel1-single_sample-10 { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-single_sample-10 { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-single_sample-10 { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-bunch_of_samples-auto[ ```r # A bunch of sample means *last_plot() ``` ] .panel2-bunch_of_samples-auto[ <!-- --> ] --- count: false .panel1-bunch_of_samples-auto[ ```r # A bunch of sample means last_plot() + * ggsample::facet_sample(n_facets = 20, n_sampled = 20) ``` ] .panel2-bunch_of_samples-auto[ <!-- --> ] <style> .panel1-bunch_of_samples-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-bunch_of_samples-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-bunch_of_samples-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-make_a_function-auto[ ```r # How can we take many samples *sample_wait_times <- function(x){ * sample(x = faithful$waiting, size = 20) *} ``` ] .panel2-make_a_function-auto[ ] --- count: false .panel1-make_a_function-auto[ ```r # How can we take many samples sample_wait_times <- function(x){ sample(x = faithful$waiting, size = 20) } # check that function is working *sample_wait_times() ``` ] .panel2-make_a_function-auto[ ``` [1] 56 89 49 75 79 64 89 82 77 74 48 85 53 45 67 82 54 58 50 82 ``` ] <style> .panel1-make_a_function-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-make_a_function-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-make_a_function-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-normal_dist-auto[ ```r *set.seed(12345) ``` ] .panel2-normal_dist-auto[ ] --- count: false .panel1-normal_dist-auto[ ```r set.seed(12345) *tibble(x = 1:10000) # for 1000 trials ``` ] .panel2-normal_dist-auto[ ``` # A tibble: 10,000 x 1 x <int> 1 1 2 2 3 3 4 4 5 5 6 6 7 7 8 8 9 9 10 10 11 11 12 12 13 13 14 14 15 15 16 16 17 17 18 18 19 19 20 20 21 21 22 22 23 23 24 24 25 25 26 26 27 27 28 28 29 29 30 30 31 31 32 32 33 33 34 34 35 35 36 36 37 37 38 38 39 39 40 40 41 41 42 42 43 43 44 44 45 45 46 46 47 47 48 48 49 49 50 50 51 51 52 52 53 53 54 54 55 55 # ... with 9,945 more rows ``` ] --- count: false .panel1-normal_dist-auto[ ```r set.seed(12345) tibble(x = 1:10000) %>% # for 1000 trials * mutate(sample = map(x, sample_wait_times)) # sample ``` ] .panel2-normal_dist-auto[ ``` # A tibble: 10,000 x 2 x sample <int> <list> 1 1 <dbl [20]> 2 2 <dbl [20]> 3 3 <dbl [20]> 4 4 <dbl [20]> 5 5 <dbl [20]> 6 6 <dbl [20]> 7 7 <dbl [20]> 8 8 <dbl [20]> 9 9 <dbl [20]> 10 10 <dbl [20]> 11 11 <dbl [20]> 12 12 <dbl [20]> 13 13 <dbl [20]> 14 14 <dbl [20]> 15 15 <dbl [20]> 16 16 <dbl [20]> 17 17 <dbl [20]> 18 18 <dbl [20]> 19 19 <dbl [20]> 20 20 <dbl [20]> 21 21 <dbl [20]> 22 22 <dbl [20]> 23 23 <dbl [20]> 24 24 <dbl [20]> 25 25 <dbl [20]> 26 26 <dbl [20]> 27 27 <dbl [20]> 28 28 <dbl [20]> 29 29 <dbl [20]> 30 30 <dbl [20]> 31 31 <dbl [20]> 32 32 <dbl [20]> 33 33 <dbl [20]> 34 34 <dbl [20]> 35 35 <dbl [20]> 36 36 <dbl [20]> 37 37 <dbl [20]> 38 38 <dbl [20]> 39 39 <dbl [20]> 40 40 <dbl [20]> 41 41 <dbl [20]> 42 42 <dbl [20]> 43 43 <dbl [20]> 44 44 <dbl [20]> 45 45 <dbl [20]> 46 46 <dbl [20]> 47 47 <dbl [20]> 48 48 <dbl [20]> 49 49 <dbl [20]> 50 50 <dbl [20]> 51 51 <dbl [20]> 52 52 <dbl [20]> 53 53 <dbl [20]> 54 54 <dbl [20]> 55 55 <dbl [20]> # ... with 9,945 more rows ``` ] --- count: false .panel1-normal_dist-auto[ ```r set.seed(12345) tibble(x = 1:10000) %>% # for 1000 trials mutate(sample = map(x, sample_wait_times)) %>% # sample * mutate(sample_mean = map_dbl(sample, mean)) # and compute means ``` ] .panel2-normal_dist-auto[ ``` # A tibble: 10,000 x 3 x sample sample_mean <int> <list> <dbl> 1 1 <dbl [20]> 71.0 2 2 <dbl [20]> 65.4 3 3 <dbl [20]> 69.6 4 4 <dbl [20]> 76.8 5 5 <dbl [20]> 71.3 6 6 <dbl [20]> 68.7 7 7 <dbl [20]> 73.1 8 8 <dbl [20]> 77.5 9 9 <dbl [20]> 73.3 10 10 <dbl [20]> 68.9 11 11 <dbl [20]> 76.0 12 12 <dbl [20]> 68.0 13 13 <dbl [20]> 71.6 14 14 <dbl [20]> 71.4 15 15 <dbl [20]> 67.6 16 16 <dbl [20]> 67.0 17 17 <dbl [20]> 75 18 18 <dbl [20]> 71.2 19 19 <dbl [20]> 72.7 20 20 <dbl [20]> 70.4 21 21 <dbl [20]> 72.6 22 22 <dbl [20]> 69 23 23 <dbl [20]> 71.8 24 24 <dbl [20]> 70.2 25 25 <dbl [20]> 65.9 26 26 <dbl [20]> 70.5 27 27 <dbl [20]> 77.6 28 28 <dbl [20]> 70.8 29 29 <dbl [20]> 72.8 30 30 <dbl [20]> 73.6 31 31 <dbl [20]> 69.4 32 32 <dbl [20]> 75.4 33 33 <dbl [20]> 70.6 34 34 <dbl [20]> 71.8 35 35 <dbl [20]> 67.3 36 36 <dbl [20]> 68.3 37 37 <dbl [20]> 63.6 38 38 <dbl [20]> 71.4 39 39 <dbl [20]> 65.9 40 40 <dbl [20]> 75.2 41 41 <dbl [20]> 65.4 42 42 <dbl [20]> 66.2 43 43 <dbl [20]> 75.2 44 44 <dbl [20]> 71.3 45 45 <dbl [20]> 71.4 46 46 <dbl [20]> 77.3 47 47 <dbl [20]> 68.9 48 48 <dbl [20]> 71.8 49 49 <dbl [20]> 69.4 50 50 <dbl [20]> 65.2 51 51 <dbl [20]> 65.8 52 52 <dbl [20]> 76.4 53 53 <dbl [20]> 75.0 54 54 <dbl [20]> 69 55 55 <dbl [20]> 72.0 # ... with 9,945 more rows ``` ] --- count: false .panel1-normal_dist-auto[ ```r set.seed(12345) tibble(x = 1:10000) %>% # for 1000 trials mutate(sample = map(x, sample_wait_times)) %>% # sample mutate(sample_mean = map_dbl(sample, mean)) %>% # and compute means * ggplot() ``` ] .panel2-normal_dist-auto[ <!-- --> ] --- count: false .panel1-normal_dist-auto[ ```r set.seed(12345) tibble(x = 1:10000) %>% # for 1000 trials mutate(sample = map(x, sample_wait_times)) %>% # sample mutate(sample_mean = map_dbl(sample, mean)) %>% # and compute means ggplot() + * aes(x = sample_mean) ``` ] .panel2-normal_dist-auto[ <!-- --> ] --- count: false .panel1-normal_dist-auto[ ```r set.seed(12345) tibble(x = 1:10000) %>% # for 1000 trials mutate(sample = map(x, sample_wait_times)) %>% # sample mutate(sample_mean = map_dbl(sample, mean)) %>% # and compute means ggplot() + aes(x = sample_mean) + * geom_rug() ``` ] .panel2-normal_dist-auto[ <!-- --> ] --- count: false .panel1-normal_dist-auto[ ```r set.seed(12345) tibble(x = 1:10000) %>% # for 1000 trials mutate(sample = map(x, sample_wait_times)) %>% # sample mutate(sample_mean = map_dbl(sample, mean)) %>% # and compute means ggplot() + aes(x = sample_mean) + geom_rug() + * geom_histogram() ``` ] .panel2-normal_dist-auto[ <!-- --> ] --- count: false .panel1-normal_dist-auto[ ```r set.seed(12345) tibble(x = 1:10000) %>% # for 1000 trials mutate(sample = map(x, sample_wait_times)) %>% # sample mutate(sample_mean = map_dbl(sample, mean)) %>% # and compute means ggplot() + aes(x = sample_mean) + geom_rug() + geom_histogram() + * ggxmean::geom_normal_dist(fill = "magenta", * height = 7000) ``` ] .panel2-normal_dist-auto[ <!-- --> ] --- count: false .panel1-normal_dist-auto[ ```r set.seed(12345) tibble(x = 1:10000) %>% # for 1000 trials mutate(sample = map(x, sample_wait_times)) %>% # sample mutate(sample_mean = map_dbl(sample, mean)) %>% # and compute means ggplot() + aes(x = sample_mean) + geom_rug() + geom_histogram() + ggxmean::geom_normal_dist(fill = "magenta", height = 7000) + * ggxmean::geom_normal_dist_zlines(color = "magenta", * height = 7000) ``` ] .panel2-normal_dist-auto[ <!-- --> ] --- count: false .panel1-normal_dist-auto[ ```r set.seed(12345) tibble(x = 1:10000) %>% # for 1000 trials mutate(sample = map(x, sample_wait_times)) %>% # sample mutate(sample_mean = map_dbl(sample, mean)) %>% # and compute means ggplot() + aes(x = sample_mean) + geom_rug() + geom_histogram() + ggxmean::geom_normal_dist(fill = "magenta", height = 7000) + ggxmean::geom_normal_dist_zlines(color = "magenta", height = 7000) + * ggxmean::geom_x_mean() ``` ] .panel2-normal_dist-auto[ <!-- --> ] --- count: false .panel1-normal_dist-auto[ ```r set.seed(12345) tibble(x = 1:10000) %>% # for 1000 trials mutate(sample = map(x, sample_wait_times)) %>% # sample mutate(sample_mean = map_dbl(sample, mean)) %>% # and compute means ggplot() + aes(x = sample_mean) + geom_rug() + geom_histogram() + ggxmean::geom_normal_dist(fill = "magenta", height = 7000) + ggxmean::geom_normal_dist_zlines(color = "magenta", height = 7000) + ggxmean::geom_x_mean() + * ggxmean::geom_x_mean_label() ``` ] .panel2-normal_dist-auto[ <!-- --> ] <style> .panel1-normal_dist-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-normal_dist-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-normal_dist-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-faithfulsamples-auto[ ```r # And so *set.seed(123456) ``` ] .panel2-faithfulsamples-auto[ ] --- count: false .panel1-faithfulsamples-auto[ ```r # And so set.seed(123456) *ggplot(data = faithful) ``` ] .panel2-faithfulsamples-auto[ <!-- --> ] --- count: false .panel1-faithfulsamples-auto[ ```r # And so set.seed(123456) ggplot(data = faithful) + * aes(x = waiting) ``` ] .panel2-faithfulsamples-auto[ <!-- --> ] --- count: false .panel1-faithfulsamples-auto[ ```r # And so set.seed(123456) ggplot(data = faithful) + aes(x = waiting) + * geom_rug() ``` ] .panel2-faithfulsamples-auto[ <!-- --> ] --- count: false .panel1-faithfulsamples-auto[ ```r # And so set.seed(123456) ggplot(data = faithful) + aes(x = waiting) + geom_rug() + * geom_vline(xintercept = rep(71, 20), color = "red") ``` ] .panel2-faithfulsamples-auto[ <!-- --> ] --- count: false .panel1-faithfulsamples-auto[ ```r # And so set.seed(123456) ggplot(data = faithful) + aes(x = waiting) + geom_rug() + geom_vline(xintercept = rep(71, 20), color = "red") + * geom_dotplot(dotsize = .5) ``` ] .panel2-faithfulsamples-auto[ <!-- --> ] --- count: false .panel1-faithfulsamples-auto[ ```r # And so set.seed(123456) ggplot(data = faithful) + aes(x = waiting) + geom_rug() + geom_vline(xintercept = rep(71, 20), color = "red") + geom_dotplot(dotsize = .5) + * ggxmean::geom_x_mean(linetype = "dashed") ``` ] .panel2-faithfulsamples-auto[ <!-- --> ] --- count: false .panel1-faithfulsamples-auto[ ```r # And so set.seed(123456) ggplot(data = faithful) + aes(x = waiting) + geom_rug() + geom_vline(xintercept = rep(71, 20), color = "red") + geom_dotplot(dotsize = .5) + ggxmean::geom_x_mean(linetype = "dashed") + * ggxmean::geom_x_mean_label() ``` ] .panel2-faithfulsamples-auto[ <!-- --> ] --- count: false .panel1-faithfulsamples-auto[ ```r # And so set.seed(123456) ggplot(data = faithful) + aes(x = waiting) + geom_rug() + geom_vline(xintercept = rep(71, 20), color = "red") + geom_dotplot(dotsize = .5) + ggxmean::geom_x_mean(linetype = "dashed") + ggxmean::geom_x_mean_label() + * ggsample::facet_sample(n_facets = 16, * n_sampled = 20) ``` ] .panel2-faithfulsamples-auto[ <!-- --> ] --- count: false .panel1-faithfulsamples-auto[ ```r # And so set.seed(123456) ggplot(data = faithful) + aes(x = waiting) + geom_rug() + geom_vline(xintercept = rep(71, 20), color = "red") + geom_dotplot(dotsize = .5) + ggxmean::geom_x_mean(linetype = "dashed") + ggxmean::geom_x_mean_label() + ggsample::facet_sample(n_facets = 16, n_sampled = 20) + * ggxmean:::geom_tdist(height = 7, fill = "goldenrod") ``` ] .panel2-faithfulsamples-auto[ <!-- --> ] --- count: false .panel1-faithfulsamples-auto[ ```r # And so set.seed(123456) ggplot(data = faithful) + aes(x = waiting) + geom_rug() + geom_vline(xintercept = rep(71, 20), color = "red") + geom_dotplot(dotsize = .5) + ggxmean::geom_x_mean(linetype = "dashed") + ggxmean::geom_x_mean_label() + ggsample::facet_sample(n_facets = 16, n_sampled = 20) + ggxmean:::geom_tdist(height = 7, fill = "goldenrod") + * ggxmean:::geom_ttestconf(color = "darkgreen", * alpha = 1) ``` ] .panel2-faithfulsamples-auto[ <!-- --> ] <style> .panel1-faithfulsamples-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-faithfulsamples-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-faithfulsamples-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> <!-- adjust font size in this css code chunk, currently 80 --> ---