class: inverse, center, middle background-image: url(https://images.unsplash.com/photo-1509081438789-63838f150db7?ixlib=rb-4.0.3&ixid=MnwxMjA3fDB8MHxwaG90by1wYWdlfHx8fGVufDB8fHx8&auto=format&fit=crop&w=387&q=80) background-size: cover # ma206distributions and ma206equations --- class: inverse, middle, center # Hello! --- class: inverse, middle, center # Thank you for your willingess to have a look at some of the work we are doing in `ma206distributions` and `ma206equations`! --- class: inverse, middle, center # Mathematical fluency involves frequent use of probability distributions and of course equations. --- class: inverse, middle, center # However, visualizing these distributions with software and typing equations can be tedious. --- class: inverse, middle, center # In a well-defined statistics and probability curriculum, the set of distributions and equations to be mastered is known and limited. --- class: inverse, middle, center # The goal of this project is to provide ready-to-use functions that will make it easier to engage computationally with these distributions. --- class: inverse, middle, center # In what follows, we highlight and demo some of the functionality. --- class: inverse, middle, center # Your feedback is on this work is greatly appreciated. --- class: inverse, middle, center # Without further ado... <style type="text/css"> .remark-code{line-height: 1.5; font-size: 90%} </style> --- class: inverse, center, middle # tidy_dbinom() ## The function tidy_dbinom() returns a dataframe that pairs outcomes and probabilities, using just two inputs. View source code [here](https://github.com/EvaMaeRey/ma206distributions/blob/main/R/tidy_dbinom.R). --- count: false ### tidy_dbinom() usage .panel1-tidy_dbinom-auto[ ```r *library(tidyverse) ``` ] .panel2-tidy_dbinom-auto[ ] --- count: false ### tidy_dbinom() usage .panel1-tidy_dbinom-auto[ ```r library(tidyverse) *library(ma206distributions) ``` ] .panel2-tidy_dbinom-auto[ ] --- count: false ### tidy_dbinom() usage .panel1-tidy_dbinom-auto[ ```r library(tidyverse) library(ma206distributions) *library(ma206equations) ``` ] .panel2-tidy_dbinom-auto[ ] --- count: false ### tidy_dbinom() usage .panel1-tidy_dbinom-auto[ ```r library(tidyverse) library(ma206distributions) library(ma206equations) *tidy_dbinom(single_trial_prob = 1/6, * num_trials = 8) ``` ] .panel2-tidy_dbinom-auto[ ``` # A tibble: 9 × 4 num_successes probability single_trial_prob num_trials <int> <dbl> <dbl> <dbl> 1 0 0.233 0.167 8 2 1 0.372 0.167 8 3 2 0.260 0.167 8 4 3 0.104 0.167 8 5 4 0.0260 0.167 8 6 5 0.00417 0.167 8 7 6 0.000417 0.167 8 8 7 0.0000238 0.167 8 9 8 0.000000595 0.167 8 ``` ] --- count: false ### tidy_dbinom() usage .panel1-tidy_dbinom-auto[ ```r library(tidyverse) library(ma206distributions) library(ma206equations) tidy_dbinom(single_trial_prob = 1/6, num_trials = 8) -> *eight_die_rolls ``` ] .panel2-tidy_dbinom-auto[ ] --- count: false ### tidy_dbinom() usage .panel1-tidy_dbinom-auto[ ```r library(tidyverse) library(ma206distributions) library(ma206equations) tidy_dbinom(single_trial_prob = 1/6, num_trials = 8) -> eight_die_rolls *tidy_dbinom(single_trial_prob = .5, * num_trials = 10) ``` ] .panel2-tidy_dbinom-auto[ ``` # A tibble: 11 × 4 num_successes probability single_trial_prob num_trials <int> <dbl> <dbl> <dbl> 1 0 0.000977 0.5 10 2 1 0.00977 0.5 10 3 2 0.0439 0.5 10 4 3 0.117 0.5 10 5 4 0.205 0.5 10 6 5 0.246 0.5 10 7 6 0.205 0.5 10 8 7 0.117 0.5 10 9 8 0.0439 0.5 10 10 9 0.00977 0.5 10 11 10 0.000977 0.5 10 ``` ] --- count: false ### tidy_dbinom() usage .panel1-tidy_dbinom-auto[ ```r library(tidyverse) library(ma206distributions) library(ma206equations) tidy_dbinom(single_trial_prob = 1/6, num_trials = 8) -> eight_die_rolls tidy_dbinom(single_trial_prob = .5, num_trials = 10) -> *ten_coin_flips ``` ] .panel2-tidy_dbinom-auto[ ] --- count: false ### tidy_dbinom() usage .panel1-tidy_dbinom-auto[ ```r library(tidyverse) library(ma206distributions) library(ma206equations) tidy_dbinom(single_trial_prob = 1/6, num_trials = 8) -> eight_die_rolls tidy_dbinom(single_trial_prob = .5, num_trials = 10) -> ten_coin_flips *tidy_dbinom(single_trial_prob = 1/38, * num_trials = 3) ``` ] .panel2-tidy_dbinom-auto[ ``` # A tibble: 4 × 4 num_successes probability single_trial_prob num_trials <int> <dbl> <dbl> <dbl> 1 0 0.923 0.0263 3 2 1 0.0748 0.0263 3 3 2 0.00202 0.0263 3 4 3 0.0000182 0.0263 3 ``` ] --- count: false ### tidy_dbinom() usage .panel1-tidy_dbinom-auto[ ```r library(tidyverse) library(ma206distributions) library(ma206equations) tidy_dbinom(single_trial_prob = 1/6, num_trials = 8) -> eight_die_rolls tidy_dbinom(single_trial_prob = .5, num_trials = 10) -> ten_coin_flips tidy_dbinom(single_trial_prob = 1/38, num_trials = 3) -> *three_roulette_spins ``` ] .panel2-tidy_dbinom-auto[ ] --- count: false ### tidy_dbinom() usage .panel1-tidy_dbinom-auto[ ```r library(tidyverse) library(ma206distributions) library(ma206equations) tidy_dbinom(single_trial_prob = 1/6, num_trials = 8) -> eight_die_rolls tidy_dbinom(single_trial_prob = .5, num_trials = 10) -> ten_coin_flips tidy_dbinom(single_trial_prob = 1/38, num_trials = 3) -> three_roulette_spins ``` ] .panel2-tidy_dbinom-auto[ ] <style> .panel1-tidy_dbinom-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-tidy_dbinom-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-tidy_dbinom-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- class: inverse, center, middle # geom_lollipop(), scale_x_counting(), geom_fulcrum ## The dataframe that tidy_dbinom() creates can easily be fed into the popular ggplot2 visualization system. Visualization functions geom_lollipop() and scale_x_counting() help concisely visualize discrete probability visualizations. geom_fulcrum() marks the balancing point of the data - which is the expected value. --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r *library(ma206distributions) ``` ] .panel2-tidy_dbinom_plot-auto[ ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) *library(tidyverse) ``` ] .panel2-tidy_dbinom_plot-auto[ ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) *tidy_dbinom(single_trial_prob = 1/6, * num_trials = 6) ``` ] .panel2-tidy_dbinom_plot-auto[ ``` # A tibble: 7 × 4 num_successes probability single_trial_prob num_trials <int> <dbl> <dbl> <dbl> 1 0 0.335 0.167 6 2 1 0.402 0.167 6 3 2 0.201 0.167 6 4 3 0.0536 0.167 6 5 4 0.00804 0.167 6 6 5 0.000643 0.167 6 7 6 0.0000214 0.167 6 ``` ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% * ggplot() ``` ] .panel2-tidy_dbinom_plot-auto[ <!-- --> ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% ggplot() + * aes(x = num_successes, * y = probability) ``` ] .panel2-tidy_dbinom_plot-auto[ <!-- --> ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% ggplot() + aes(x = num_successes, y = probability) + * geom_lollipop() ``` ] .panel2-tidy_dbinom_plot-auto[ <!-- --> ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + * scale_x_counting() ``` ] .panel2-tidy_dbinom_plot-auto[ <!-- --> ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + scale_x_counting() -> *die_roll_viz ``` ] .panel2-tidy_dbinom_plot-auto[ ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + scale_x_counting() -> die_roll_viz *tidy_dbinom(single_trial_prob = .5, * num_trials = 10) ``` ] .panel2-tidy_dbinom_plot-auto[ ``` # A tibble: 11 × 4 num_successes probability single_trial_prob num_trials <int> <dbl> <dbl> <dbl> 1 0 0.000977 0.5 10 2 1 0.00977 0.5 10 3 2 0.0439 0.5 10 4 3 0.117 0.5 10 5 4 0.205 0.5 10 6 5 0.246 0.5 10 7 6 0.205 0.5 10 8 7 0.117 0.5 10 9 8 0.0439 0.5 10 10 9 0.00977 0.5 10 11 10 0.000977 0.5 10 ``` ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + scale_x_counting() -> die_roll_viz tidy_dbinom(single_trial_prob = .5, num_trials = 10) %>% * ggplot() ``` ] .panel2-tidy_dbinom_plot-auto[ <!-- --> ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + scale_x_counting() -> die_roll_viz tidy_dbinom(single_trial_prob = .5, num_trials = 10) %>% ggplot() + * aes(x = num_successes, * y = probability) ``` ] .panel2-tidy_dbinom_plot-auto[ <!-- --> ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + scale_x_counting() -> die_roll_viz tidy_dbinom(single_trial_prob = .5, num_trials = 10) %>% ggplot() + aes(x = num_successes, y = probability) + * geom_lollipop() ``` ] .panel2-tidy_dbinom_plot-auto[ 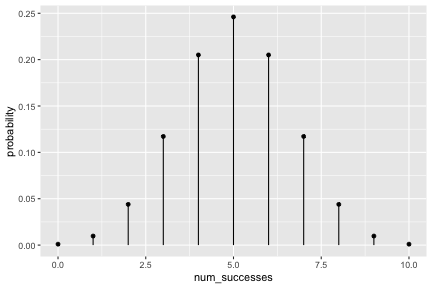<!-- --> ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + scale_x_counting() -> die_roll_viz tidy_dbinom(single_trial_prob = .5, num_trials = 10) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + * scale_x_counting() ``` ] .panel2-tidy_dbinom_plot-auto[ 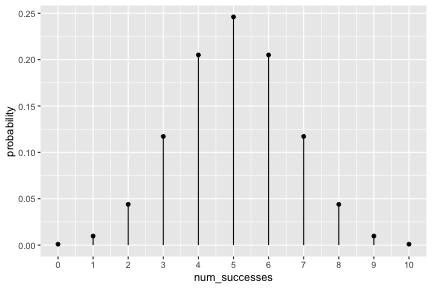<!-- --> ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + scale_x_counting() -> die_roll_viz tidy_dbinom(single_trial_prob = .5, num_trials = 10) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + scale_x_counting() -> *coin_flip_viz ``` ] .panel2-tidy_dbinom_plot-auto[ ] --- count: false ### geom_lollipop() and scale_x_counting() usage .panel1-tidy_dbinom_plot-auto[ ```r library(ma206distributions) library(tidyverse) tidy_dbinom(single_trial_prob = 1/6, num_trials = 6) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + scale_x_counting() -> die_roll_viz tidy_dbinom(single_trial_prob = .5, num_trials = 10) %>% ggplot() + aes(x = num_successes, y = probability) + geom_lollipop() + scale_x_counting() -> coin_flip_viz ``` ] .panel2-tidy_dbinom_plot-auto[ ] <style> .panel1-tidy_dbinom_plot-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-tidy_dbinom_plot-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-tidy_dbinom_plot-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- class: inverse, center, middle # stamp_dbinom() ## For convenience stamp_dbinom() wraps up the dataframe together with the plotting functionality. --- count: false ### stamp_dbinom() usage .panel1-tidy_dbinom_stamp-auto[ ```r *ggplot() ``` ] .panel2-tidy_dbinom_stamp-auto[ <!-- --> ] --- count: false ### stamp_dbinom() usage .panel1-tidy_dbinom_stamp-auto[ ```r ggplot() + * labs(title = "Probability of rolling 0,1,2,...10 sixes\nwhen rolling a fair die 10 times") ``` ] .panel2-tidy_dbinom_stamp-auto[ <!-- --> ] --- count: false ### stamp_dbinom() usage .panel1-tidy_dbinom_stamp-auto[ ```r ggplot() + labs(title = "Probability of rolling 0,1,2,...10 sixes\nwhen rolling a fair die 10 times") + * stamp_dbinom(single_trial_prob = 1/6, * num_trials = 10) ``` ] .panel2-tidy_dbinom_stamp-auto[ 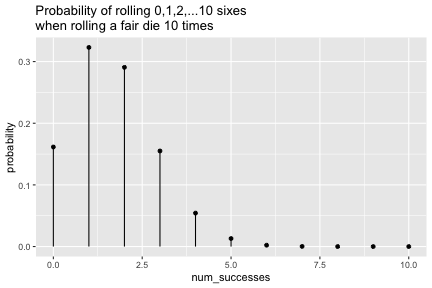<!-- --> ] --- count: false ### stamp_dbinom() usage .panel1-tidy_dbinom_stamp-auto[ ```r ggplot() + labs(title = "Probability of rolling 0,1,2,...10 sixes\nwhen rolling a fair die 10 times") + stamp_dbinom(single_trial_prob = 1/6, num_trials = 10) + * scale_x_counting() ``` ] .panel2-tidy_dbinom_stamp-auto[ 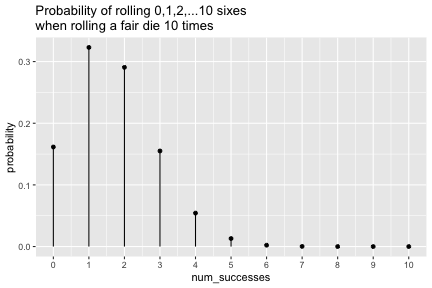<!-- --> ] --- count: false ### stamp_dbinom() usage .panel1-tidy_dbinom_stamp-auto[ ```r ggplot() + labs(title = "Probability of rolling 0,1,2,...10 sixes\nwhen rolling a fair die 10 times") + stamp_dbinom(single_trial_prob = 1/6, num_trials = 10) + scale_x_counting() -> *die_roll_viz_w_stamp ``` ] .panel2-tidy_dbinom_stamp-auto[ ] --- count: false ### stamp_dbinom() usage .panel1-tidy_dbinom_stamp-auto[ ```r ggplot() + labs(title = "Probability of rolling 0,1,2,...10 sixes\nwhen rolling a fair die 10 times") + stamp_dbinom(single_trial_prob = 1/6, num_trials = 10) + scale_x_counting() -> die_roll_viz_w_stamp *ggplot() ``` ] .panel2-tidy_dbinom_stamp-auto[ <!-- --> ] --- count: false ### stamp_dbinom() usage .panel1-tidy_dbinom_stamp-auto[ ```r ggplot() + labs(title = "Probability of rolling 0,1,2,...10 sixes\nwhen rolling a fair die 10 times") + stamp_dbinom(single_trial_prob = 1/6, num_trials = 10) + scale_x_counting() -> die_roll_viz_w_stamp ggplot() + * labs(title = "Probability of flipping 0,1,2,3,4 or 5 heads\n when flipping a coin five times") ``` ] .panel2-tidy_dbinom_stamp-auto[ <!-- --> ] --- count: false ### stamp_dbinom() usage .panel1-tidy_dbinom_stamp-auto[ ```r ggplot() + labs(title = "Probability of rolling 0,1,2,...10 sixes\nwhen rolling a fair die 10 times") + stamp_dbinom(single_trial_prob = 1/6, num_trials = 10) + scale_x_counting() -> die_roll_viz_w_stamp ggplot() + labs(title = "Probability of flipping 0,1,2,3,4 or 5 heads\n when flipping a coin five times") + * stamp_dbinom(single_trial_prob = .5, * num_trials = 5) ``` ] .panel2-tidy_dbinom_stamp-auto[ <!-- --> ] --- count: false ### stamp_dbinom() usage .panel1-tidy_dbinom_stamp-auto[ ```r ggplot() + labs(title = "Probability of rolling 0,1,2,...10 sixes\nwhen rolling a fair die 10 times") + stamp_dbinom(single_trial_prob = 1/6, num_trials = 10) + scale_x_counting() -> die_roll_viz_w_stamp ggplot() + labs(title = "Probability of flipping 0,1,2,3,4 or 5 heads\n when flipping a coin five times") + stamp_dbinom(single_trial_prob = .5, num_trials = 5) -> *coin_flip_viz_w_stamp ``` ] .panel2-tidy_dbinom_stamp-auto[ ] --- count: false ### stamp_dbinom() usage .panel1-tidy_dbinom_stamp-auto[ ```r ggplot() + labs(title = "Probability of rolling 0,1,2,...10 sixes\nwhen rolling a fair die 10 times") + stamp_dbinom(single_trial_prob = 1/6, num_trials = 10) + scale_x_counting() -> die_roll_viz_w_stamp ggplot() + labs(title = "Probability of flipping 0,1,2,3,4 or 5 heads\n when flipping a coin five times") + stamp_dbinom(single_trial_prob = .5, num_trials = 5) -> coin_flip_viz_w_stamp ``` ] .panel2-tidy_dbinom_stamp-auto[ ] <style> .panel1-tidy_dbinom_stamp-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-tidy_dbinom_stamp-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-tidy_dbinom_stamp-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- class: inverse, center, middle # stamp_eq_binomial() ## The function stamp_eq_binomial() tells you how the probability for each number of total successes is computed. --- count: false .panel1-equations-auto[ ```r *library(ma206equations) ``` ] .panel2-equations-auto[ ] --- count: false .panel1-equations-auto[ ```r library(ma206equations) *library(ma206distributions) ``` ] .panel2-equations-auto[ ] --- count: false .panel1-equations-auto[ ```r library(ma206equations) library(ma206distributions) *ggplot() ``` ] .panel2-equations-auto[ <!-- --> ] --- count: false .panel1-equations-auto[ ```r library(ma206equations) library(ma206distributions) ggplot() + * stamp_dbinom(single_trial_prob = .5, * num_trials = 5) ``` ] .panel2-equations-auto[ 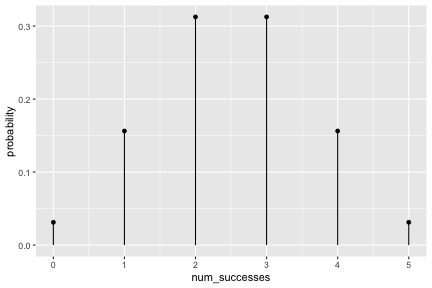<!-- --> ] --- count: false .panel1-equations-auto[ ```r library(ma206equations) library(ma206distributions) ggplot() + stamp_dbinom(single_trial_prob = .5, num_trials = 5) + # binomial computation * stamp_eq_binomial(x = 4, y = .27, size = 7) ``` ] .panel2-equations-auto[ 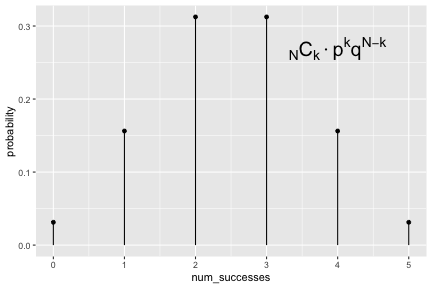<!-- --> ] --- count: false .panel1-equations-auto[ ```r library(ma206equations) library(ma206distributions) ggplot() + stamp_dbinom(single_trial_prob = .5, num_trials = 5) + # binomial computation stamp_eq_binomial(x = 4, y = .27, size = 7) + # reminder of computing choose * stamp_eq_choose(x = 4.5, y = .21) ``` ] .panel2-equations-auto[ 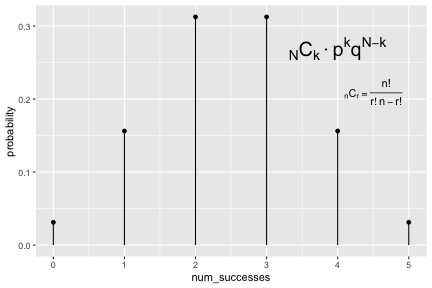<!-- --> ] <style> .panel1-equations-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-equations-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-equations-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- # To add, equations outside of context plot context. --- class: inverse, center, middle # bernoulli_trial() and adding trials() ## You can also build up binomial distributions from first principles uising bernoulli_trial() and adding trials(). --- count: false ## bernoulli_trial() and add_trials() usage .panel1-pipeline2-auto[ ```r *bernoulli_trial(prob = .25) ``` ] .panel2-pipeline2-auto[ ``` # A tibble: 2 × 2 outcome prob <int> <dbl> 1 0 0.75 2 1 0.25 ``` ] --- count: false ## bernoulli_trial() and add_trials() usage .panel1-pipeline2-auto[ ```r bernoulli_trial(prob = .25) %>% * add_trials() ``` ] .panel2-pipeline2-auto[ ``` # A tibble: 4 × 4 t1_outcome t1_prob t2_outcome t2_prob <int> <dbl> <int> <dbl> 1 0 0.75 0 0.75 2 0 0.75 1 0.25 3 1 0.25 0 0.75 4 1 0.25 1 0.25 ``` ] --- count: false ## bernoulli_trial() and add_trials() usage .panel1-pipeline2-auto[ ```r bernoulli_trial(prob = .25) %>% add_trials() %>% * add_trials() ``` ] .panel2-pipeline2-auto[ ``` # A tibble: 8 × 6 t1_outcome t1_prob t2_outcome t2_prob t3_outcome t3_prob <int> <dbl> <int> <dbl> <int> <dbl> 1 0 0.75 0 0.75 0 0.75 2 0 0.75 0 0.75 1 0.25 3 0 0.75 1 0.25 0 0.75 4 0 0.75 1 0.25 1 0.25 5 1 0.25 0 0.75 0 0.75 6 1 0.25 0 0.75 1 0.25 7 1 0.25 1 0.25 0 0.75 8 1 0.25 1 0.25 1 0.25 ``` ] --- count: false ## bernoulli_trial() and add_trials() usage .panel1-pipeline2-auto[ ```r bernoulli_trial(prob = .25) %>% add_trials() %>% add_trials() %>% * .$out ``` ] .panel2-pipeline2-auto[ ``` # A tibble: 8 × 6 t1_outcome t1_prob t2_outcome t2_prob t3_outcome t3_prob <int> <dbl> <int> <dbl> <int> <dbl> 1 0 0.75 0 0.75 0 0.75 2 0 0.75 0 0.75 1 0.25 3 0 0.75 1 0.25 0 0.75 4 0 0.75 1 0.25 1 0.25 5 1 0.25 0 0.75 0 0.75 6 1 0.25 0 0.75 1 0.25 7 1 0.25 1 0.25 0 0.75 8 1 0.25 1 0.25 1 0.25 ``` ] --- count: false ## bernoulli_trial() and add_trials() usage .panel1-pipeline2-auto[ ```r bernoulli_trial(prob = .25) %>% add_trials() %>% add_trials() %>% .$out %>% * sum_across() ``` ] .panel2-pipeline2-auto[ ``` # A tibble: 8 × 7 t1_outcome t1_prob t2_outcome t2_prob t3_outcome t3_prob global_outcome <int> <dbl> <int> <dbl> <int> <dbl> <dbl> 1 0 0.75 0 0.75 0 0.75 0 2 0 0.75 0 0.75 1 0.25 1 3 0 0.75 1 0.25 0 0.75 1 4 0 0.75 1 0.25 1 0.25 2 5 1 0.25 0 0.75 0 0.75 1 6 1 0.25 0 0.75 1 0.25 2 7 1 0.25 1 0.25 0 0.75 2 8 1 0.25 1 0.25 1 0.25 3 ``` ] --- count: false ## bernoulli_trial() and add_trials() usage .panel1-pipeline2-auto[ ```r bernoulli_trial(prob = .25) %>% add_trials() %>% add_trials() %>% .$out %>% sum_across() %>% * prod_across() ``` ] .panel2-pipeline2-auto[ ``` # A tibble: 8 × 8 t1_outcome t1_prob t2_outcome t2_prob t3_outcome t3_prob global_outcome global_probs <int> <dbl> <int> <dbl> <int> <dbl> <dbl> <dbl> 1 0 0.75 0 0.75 0 0.75 0 0.422 2 0 0.75 0 0.75 1 0.25 1 0.141 3 0 0.75 1 0.25 0 0.75 1 0.141 4 0 0.75 1 0.25 1 0.25 2 0.0469 5 1 0.25 0 0.75 0 0.75 1 0.141 6 1 0.25 0 0.75 1 0.25 2 0.0469 7 1 0.25 1 0.25 0 0.75 2 0.0469 8 1 0.25 1 0.25 1 0.25 3 0.0156 ``` ] --- count: false ## bernoulli_trial() and add_trials() usage .panel1-pipeline2-auto[ ```r bernoulli_trial(prob = .25) %>% add_trials() %>% add_trials() %>% .$out %>% sum_across() %>% prod_across() %>% * group_by(global_outcome) ``` ] .panel2-pipeline2-auto[ ``` # A tibble: 8 × 8 # Groups: global_outcome [4] t1_outcome t1_prob t2_outcome t2_prob t3_outcome t3_prob global_outcome global_probs <int> <dbl> <int> <dbl> <int> <dbl> <dbl> <dbl> 1 0 0.75 0 0.75 0 0.75 0 0.422 2 0 0.75 0 0.75 1 0.25 1 0.141 3 0 0.75 1 0.25 0 0.75 1 0.141 4 0 0.75 1 0.25 1 0.25 2 0.0469 5 1 0.25 0 0.75 0 0.75 1 0.141 6 1 0.25 0 0.75 1 0.25 2 0.0469 7 1 0.25 1 0.25 0 0.75 2 0.0469 8 1 0.25 1 0.25 1 0.25 3 0.0156 ``` ] --- count: false ## bernoulli_trial() and add_trials() usage .panel1-pipeline2-auto[ ```r bernoulli_trial(prob = .25) %>% add_trials() %>% add_trials() %>% .$out %>% sum_across() %>% prod_across() %>% group_by(global_outcome) %>% * summarize(probs = * sum(global_probs)) ``` ] .panel2-pipeline2-auto[ ``` # A tibble: 4 × 2 global_outcome probs <dbl> <dbl> 1 0 0.422 2 1 0.422 3 2 0.141 4 3 0.0156 ``` ] --- count: false ## bernoulli_trial() and add_trials() usage .panel1-pipeline2-auto[ ```r bernoulli_trial(prob = .25) %>% add_trials() %>% add_trials() %>% .$out %>% sum_across() %>% prod_across() %>% group_by(global_outcome) %>% summarize(probs = sum(global_probs)) # check consistency with # our automated computation *tidy_dbinom( * single_trial_pro = .25, * num_trials = 3) ``` ] .panel2-pipeline2-auto[ ``` # A tibble: 4 × 2 global_outcome probs <dbl> <dbl> 1 0 0.422 2 1 0.422 3 2 0.141 4 3 0.0156 ``` ``` # A tibble: 4 × 4 num_successes probability single_trial_prob num_trials <int> <dbl> <dbl> <dbl> 1 0 0.422 0.25 3 2 1 0.422 0.25 3 3 2 0.141 0.25 3 4 3 0.0156 0.25 3 ``` ] <style> .panel1-pipeline2-auto { color: black; width: 28%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-pipeline2-auto { color: black; width: 70%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-pipeline2-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style>