count: false .panel1-the_chunk-auto[ ``` r *library(ggswim) ``` ] .panel2-the_chunk-auto[ ] --- count: false .panel1-the_chunk-auto[ ``` r library(ggswim) *library(ggplot2) ``` ] .panel2-the_chunk-auto[ ] --- count: false .panel1-the_chunk-auto[ ``` r library(ggswim) library(ggplot2) *patient_data %>% head(3) ``` ] .panel2-the_chunk-auto[ ``` # A tibble: 3 × 4 pt_id disease_assessment start_time end_time <chr> <chr> <dbl> <dbl> 1 01 CR/CRi + B Cell Recovery -2.8 0 2 01 RD 0 0.9 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 ``` ] --- count: false .panel1-the_chunk-auto[ ``` r library(ggswim) library(ggplot2) patient_data %>% head(3) *ggplot(data = patient_data) ``` ] .panel2-the_chunk-auto[ ``` # A tibble: 3 × 4 pt_id disease_assessment start_time end_time <chr> <chr> <dbl> <dbl> 1 01 CR/CRi + B Cell Recovery -2.8 0 2 01 RD 0 0.9 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 ``` <!-- --> ] --- count: false .panel1-the_chunk-auto[ ``` r library(ggswim) library(ggplot2) patient_data %>% head(3) ggplot(data = patient_data) + * aes(x = start_time, * y = pt_id) ``` ] .panel2-the_chunk-auto[ ``` # A tibble: 3 × 4 pt_id disease_assessment start_time end_time <chr> <chr> <dbl> <dbl> 1 01 CR/CRi + B Cell Recovery -2.8 0 2 01 RD 0 0.9 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 ``` <!-- --> ] --- count: false .panel1-the_chunk-auto[ ``` r library(ggswim) library(ggplot2) patient_data %>% head(3) ggplot(data = patient_data) + aes(x = start_time, y = pt_id) + * labs(title = "My Swimmer Plot") ``` ] .panel2-the_chunk-auto[ ``` # A tibble: 3 × 4 pt_id disease_assessment start_time end_time <chr> <chr> <dbl> <dbl> 1 01 CR/CRi + B Cell Recovery -2.8 0 2 01 RD 0 0.9 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 ``` <!-- --> ] --- count: false .panel1-the_chunk-auto[ ``` r library(ggswim) library(ggplot2) patient_data %>% head(3) ggplot(data = patient_data) + aes(x = start_time, y = pt_id) + labs(title = "My Swimmer Plot") + * labs(x = "Time Since Infusion (Months)") ``` ] .panel2-the_chunk-auto[ ``` # A tibble: 3 × 4 pt_id disease_assessment start_time end_time <chr> <chr> <dbl> <dbl> 1 01 CR/CRi + B Cell Recovery -2.8 0 2 01 RD 0 0.9 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 ``` <!-- --> ] --- count: false .panel1-the_chunk-auto[ ``` r library(ggswim) library(ggplot2) patient_data %>% head(3) ggplot(data = patient_data) + aes(x = start_time, y = pt_id) + labs(title = "My Swimmer Plot") + labs(x = "Time Since Infusion (Months)") + * labs(y = "Patient ID") ``` ] .panel2-the_chunk-auto[ ``` # A tibble: 3 × 4 pt_id disease_assessment start_time end_time <chr> <chr> <dbl> <dbl> 1 01 CR/CRi + B Cell Recovery -2.8 0 2 01 RD 0 0.9 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 ``` <!-- --> ] --- count: false .panel1-the_chunk-auto[ ``` r library(ggswim) library(ggplot2) patient_data %>% head(3) ggplot(data = patient_data) + aes(x = start_time, y = pt_id) + labs(title = "My Swimmer Plot") + labs(x = "Time Since Infusion (Months)") + labs(y = "Patient ID") + * theme_ggswim() ``` ] .panel2-the_chunk-auto[ ``` # A tibble: 3 × 4 pt_id disease_assessment start_time end_time <chr> <chr> <dbl> <dbl> 1 01 CR/CRi + B Cell Recovery -2.8 0 2 01 RD 0 0.9 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 ``` <!-- --> ] --- count: false .panel1-the_chunk-auto[ ``` r library(ggswim) library(ggplot2) patient_data %>% head(3) ggplot(data = patient_data) + aes(x = start_time, y = pt_id) + labs(title = "My Swimmer Plot") + labs(x = "Time Since Infusion (Months)") + labs(y = "Patient ID") + theme_ggswim() + * geom_swim_lane(aes( * x = start_time, * y = pt_id, * xend = end_time, * color = disease_assessment)) ``` ] .panel2-the_chunk-auto[ ``` # A tibble: 3 × 4 pt_id disease_assessment start_time end_time <chr> <chr> <dbl> <dbl> 1 01 CR/CRi + B Cell Recovery -2.8 0 2 01 RD 0 0.9 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 ``` 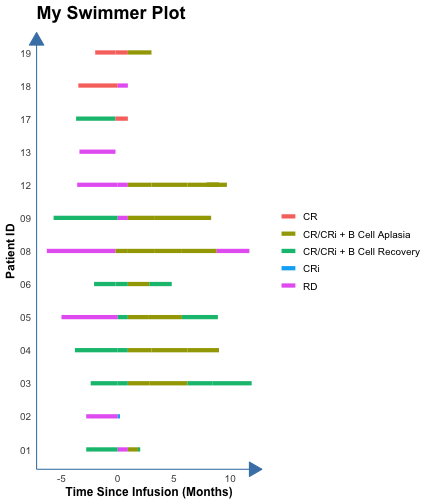<!-- --> ] --- count: false .panel1-the_chunk-auto[ ``` r library(ggswim) library(ggplot2) patient_data %>% head(3) ggplot(data = patient_data) + aes(x = start_time, y = pt_id) + labs(title = "My Swimmer Plot") + labs(x = "Time Since Infusion (Months)") + labs(y = "Patient ID") + theme_ggswim() + geom_swim_lane(aes( x = start_time, y = pt_id, xend = end_time, color = disease_assessment)) + * scale_color_brewer( * name = "Overall Disease Assessment", * palette = "Set1" * ) ``` ] .panel2-the_chunk-auto[ ``` # A tibble: 3 × 4 pt_id disease_assessment start_time end_time <chr> <chr> <dbl> <dbl> 1 01 CR/CRi + B Cell Recovery -2.8 0 2 01 RD 0 0.9 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 ``` 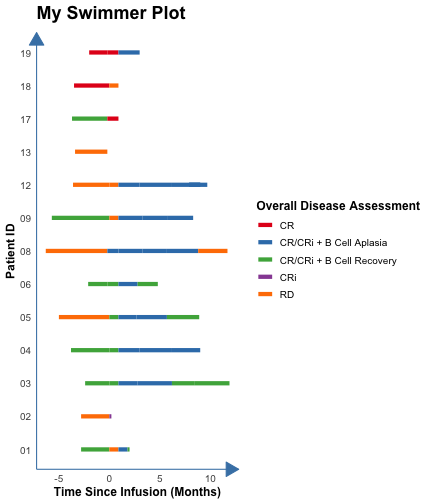<!-- --> ] --- count: false .panel1-the_chunk-auto[ ``` r library(ggswim) library(ggplot2) patient_data %>% head(3) ggplot(data = patient_data) + aes(x = start_time, y = pt_id) + labs(title = "My Swimmer Plot") + labs(x = "Time Since Infusion (Months)") + labs(y = "Patient ID") + theme_ggswim() + geom_swim_lane(aes( x = start_time, y = pt_id, xend = end_time, color = disease_assessment)) + scale_color_brewer( name = "Overall Disease Assessment", palette = "Set1" ) -> *p1 ``` ] .panel2-the_chunk-auto[ ``` # A tibble: 3 × 4 pt_id disease_assessment start_time end_time <chr> <chr> <dbl> <dbl> 1 01 CR/CRi + B Cell Recovery -2.8 0 2 01 RD 0 0.9 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 ``` ] <style> .panel1-the_chunk-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-the_chunk-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-the_chunk-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-the_chunk2-auto[ ``` r *patient_data ``` ] .panel2-the_chunk2-auto[ ``` # A tibble: 53 × 4 pt_id disease_assessment start_time end_time <chr> <chr> <dbl> <dbl> 1 01 CR/CRi + B Cell Recovery -2.8 0 2 01 RD 0 0.9 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 4 01 CR/CRi + B Cell Recovery 1.8 2 5 02 RD -2.8 0 6 02 CRi 0 0.2 7 03 CR/CRi + B Cell Recovery -2.4 0 8 03 CR/CRi + B Cell Recovery 0 0.9 9 03 CR/CRi + B Cell Aplasia 0.9 2.8 10 03 CR/CRi + B Cell Aplasia 2.8 6.2 # ℹ 43 more rows ``` ] --- count: false .panel1-the_chunk2-auto[ ``` r patient_data |> * dplyr::left_join( * end_study_events |> * dplyr::select(pt_id, end_study_name), * by = "pt_id" * ) ``` ] .panel2-the_chunk2-auto[ ``` # A tibble: 53 × 5 pt_id disease_assessment start_time end_time end_study_name <chr> <chr> <dbl> <dbl> <chr> 1 01 CR/CRi + B Cell Recovery -2.8 0 Other End Study Reason 2 01 RD 0 0.9 Other End Study Reason 3 01 CR/CRi + B Cell Aplasia 0.9 1.8 Other End Study Reason 4 01 CR/CRi + B Cell Recovery 1.8 2 Other End Study Reason 5 02 RD -2.8 0 Deceased 6 02 CRi 0 0.2 Deceased 7 03 CR/CRi + B Cell Recovery -2.4 0 Completed Study Follow-Up 8 03 CR/CRi + B Cell Recovery 0 0.9 Completed Study Follow-Up 9 03 CR/CRi + B Cell Aplasia 0.9 2.8 Completed Study Follow-Up 10 03 CR/CRi + B Cell Aplasia 2.8 6.2 Completed Study Follow-Up # ℹ 43 more rows ``` ] --- count: false .panel1-the_chunk2-auto[ ``` r patient_data |> dplyr::left_join( end_study_events |> dplyr::select(pt_id, end_study_name), by = "pt_id" ) |> * dplyr::select(pt_id, end_time, end_study_name) ``` ] .panel2-the_chunk2-auto[ ``` # A tibble: 53 × 3 pt_id end_time end_study_name <chr> <dbl> <chr> 1 01 0 Other End Study Reason 2 01 0.9 Other End Study Reason 3 01 1.8 Other End Study Reason 4 01 2 Other End Study Reason 5 02 0 Deceased 6 02 0.2 Deceased 7 03 0 Completed Study Follow-Up 8 03 0.9 Completed Study Follow-Up 9 03 2.8 Completed Study Follow-Up 10 03 6.2 Completed Study Follow-Up # ℹ 43 more rows ``` ] --- count: false .panel1-the_chunk2-auto[ ``` r patient_data |> dplyr::left_join( end_study_events |> dplyr::select(pt_id, end_study_name), by = "pt_id" ) |> dplyr::select(pt_id, end_time, end_study_name) |> * dplyr::filter(.by = pt_id, * end_time == max(end_time)) ``` ] .panel2-the_chunk2-auto[ ``` # A tibble: 14 × 3 pt_id end_time end_study_name <chr> <dbl> <chr> 1 01 2 Other End Study Reason 2 02 0.2 Deceased 3 03 11.9 Completed Study Follow-Up 4 03 11.9 Completed Study Follow-Up 5 04 9 <NA> 6 05 8.9 <NA> 7 06 4.8 Other End Study Reason 8 08 11.7 Other End Study Reason 9 09 8.3 <NA> 10 12 9.7 Other End Study Reason 11 13 -0.2 <NA> 12 17 0.9 <NA> 13 18 0.9 <NA> 14 19 3 <NA> ``` ] --- count: false .panel1-the_chunk2-auto[ ``` r patient_data |> dplyr::left_join( end_study_events |> dplyr::select(pt_id, end_study_name), by = "pt_id" ) |> dplyr::select(pt_id, end_time, end_study_name) |> dplyr::filter(.by = pt_id, end_time == max(end_time)) |> * unique() ``` ] .panel2-the_chunk2-auto[ ``` # A tibble: 13 × 3 pt_id end_time end_study_name <chr> <dbl> <chr> 1 01 2 Other End Study Reason 2 02 0.2 Deceased 3 03 11.9 Completed Study Follow-Up 4 04 9 <NA> 5 05 8.9 <NA> 6 06 4.8 Other End Study Reason 7 08 11.7 Other End Study Reason 8 09 8.3 <NA> 9 12 9.7 Other End Study Reason 10 13 -0.2 <NA> 11 17 0.9 <NA> 12 18 0.9 <NA> 13 19 3 <NA> ``` ] --- count: false .panel1-the_chunk2-auto[ ``` r patient_data |> dplyr::left_join( end_study_events |> dplyr::select(pt_id, end_study_name), by = "pt_id" ) |> dplyr::select(pt_id, end_time, end_study_name) |> dplyr::filter(.by = pt_id, end_time == max(end_time)) |> unique() -> *arrow_data ``` ] .panel2-the_chunk2-auto[ ] --- count: false .panel1-the_chunk2-auto[ ``` r patient_data |> dplyr::left_join( end_study_events |> dplyr::select(pt_id, end_study_name), by = "pt_id" ) |> dplyr::select(pt_id, end_time, end_study_name) |> dplyr::filter(.by = pt_id, end_time == max(end_time)) |> unique() -> arrow_data *arrow_data %>% head(3) ``` ] .panel2-the_chunk2-auto[ ``` # A tibble: 3 × 3 pt_id end_time end_study_name <chr> <dbl> <chr> 1 01 2 Other End Study Reason 2 02 0.2 Deceased 3 03 11.9 Completed Study Follow-Up ``` ] --- count: false .panel1-the_chunk2-auto[ ``` r patient_data |> dplyr::left_join( end_study_events |> dplyr::select(pt_id, end_study_name), by = "pt_id" ) |> dplyr::select(pt_id, end_time, end_study_name) |> dplyr::filter(.by = pt_id, end_time == max(end_time)) |> unique() -> arrow_data arrow_data %>% head(3) *p1 ``` ] .panel2-the_chunk2-auto[ ``` # A tibble: 3 × 3 pt_id end_time end_study_name <chr> <dbl> <chr> 1 01 2 Other End Study Reason 2 02 0.2 Deceased 3 03 11.9 Completed Study Follow-Up ``` 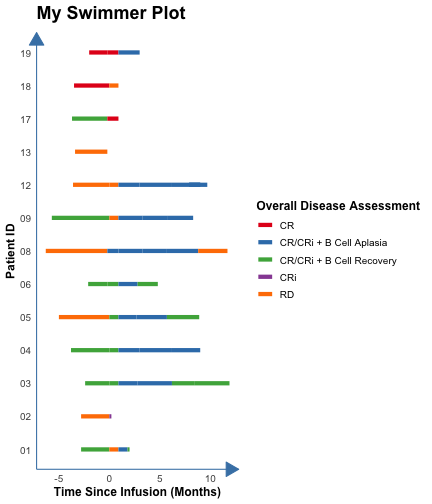<!-- --> ] --- count: false .panel1-the_chunk2-auto[ ``` r patient_data |> dplyr::left_join( end_study_events |> dplyr::select(pt_id, end_study_name), by = "pt_id" ) |> dplyr::select(pt_id, end_time, end_study_name) |> dplyr::filter(.by = pt_id, end_time == max(end_time)) |> unique() -> arrow_data arrow_data %>% head(3) p1 + * geom_point(data = arrow_data, * aes(x = end_time, * y = pt_id)) ``` ] .panel2-the_chunk2-auto[ ``` # A tibble: 3 × 3 pt_id end_time end_study_name <chr> <dbl> <chr> 1 01 2 Other End Study Reason 2 02 0.2 Deceased 3 03 11.9 Completed Study Follow-Up ``` <!-- --> ] --- count: false .panel1-the_chunk2-auto[ ``` r patient_data |> dplyr::left_join( end_study_events |> dplyr::select(pt_id, end_study_name), by = "pt_id" ) |> dplyr::select(pt_id, end_time, end_study_name) |> dplyr::filter(.by = pt_id, end_time == max(end_time)) |> unique() -> arrow_data arrow_data %>% head(3) p1 + geom_point(data = arrow_data, aes(x = end_time, y = pt_id)) + * geom_swim_arrow( * data = arrow_data, * mapping = aes(x = NULL, * y = pt_id, * xend = end_time), * size = .3) ``` ] .panel2-the_chunk2-auto[ ``` # A tibble: 3 × 3 pt_id end_time end_study_name <chr> <dbl> <chr> 1 01 2 Other End Study Reason 2 02 0.2 Deceased 3 03 11.9 Completed Study Follow-Up ``` <!-- --> ] --- count: false .panel1-the_chunk2-auto[ ``` r patient_data |> dplyr::left_join( end_study_events |> dplyr::select(pt_id, end_study_name), by = "pt_id" ) |> dplyr::select(pt_id, end_time, end_study_name) |> dplyr::filter(.by = pt_id, end_time == max(end_time)) |> unique() -> arrow_data arrow_data %>% head(3) p1 + geom_point(data = arrow_data, aes(x = end_time, y = pt_id)) + geom_swim_arrow( data = arrow_data, mapping = aes(x = NULL, y = pt_id, xend = end_time), size = .3) -> *p2 ``` ] .panel2-the_chunk2-auto[ ``` # A tibble: 3 × 3 pt_id end_time end_study_name <chr> <dbl> <chr> 1 01 2 Other End Study Reason 2 02 0.2 Deceased 3 03 11.9 Completed Study Follow-Up ``` ] <style> .panel1-the_chunk2-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-the_chunk2-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-the_chunk2-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-the_chunk3-auto[ ``` r *infusion_events %>% head(2) ``` ] .panel2-the_chunk3-auto[ ``` # A tibble: 2 × 3 pt_id time_from_initial_infusion infusion_type <chr> <dbl> <chr> 1 01 0 Initial Infusion 2 02 0 Initial Infusion ``` ] --- count: false .panel1-the_chunk3-auto[ ``` r infusion_events %>% head(2) *p2 ``` ] .panel2-the_chunk3-auto[ ``` # A tibble: 2 × 3 pt_id time_from_initial_infusion infusion_type <chr> <dbl> <chr> 1 01 0 Initial Infusion 2 02 0 Initial Infusion ``` <!-- --> ] --- count: false .panel1-the_chunk3-auto[ ``` r infusion_events %>% head(2) p2 + * new_scale_color() ``` ] .panel2-the_chunk3-auto[ ``` # A tibble: 2 × 3 pt_id time_from_initial_infusion infusion_type <chr> <dbl> <chr> 1 01 0 Initial Infusion 2 02 0 Initial Infusion ``` <!-- --> ] --- count: false .panel1-the_chunk3-auto[ ``` r infusion_events %>% head(2) p2 + new_scale_color() + * geom_point(data = infusion_events, * aes(x = time_from_initial_infusion, * y = pt_id, * color = infusion_type), * size = 5) ``` ] .panel2-the_chunk3-auto[ ``` # A tibble: 2 × 3 pt_id time_from_initial_infusion infusion_type <chr> <dbl> <chr> 1 01 0 Initial Infusion 2 02 0 Initial Infusion ``` <!-- --> ] --- count: false .panel1-the_chunk3-auto[ ``` r infusion_events %>% head(2) p2 + new_scale_color() + geom_point(data = infusion_events, aes(x = time_from_initial_infusion, y = pt_id, color = infusion_type), size = 5) + * geom_swim_point( * data = infusion_events, * aes(x = time_from_initial_infusion, * y = pt_id, * color = infusion_type), * size = 5 * ) ``` ] .panel2-the_chunk3-auto[ ``` # A tibble: 2 × 3 pt_id time_from_initial_infusion infusion_type <chr> <dbl> <chr> 1 01 0 Initial Infusion 2 02 0 Initial Infusion ``` <!-- --> ] --- count: false .panel1-the_chunk3-auto[ ``` r infusion_events %>% head(2) p2 + new_scale_color() + geom_point(data = infusion_events, aes(x = time_from_initial_infusion, y = pt_id, color = infusion_type), size = 5) + geom_swim_point( data = infusion_events, aes(x = time_from_initial_infusion, y = pt_id, color = infusion_type), size = 5 ) -> *p3 ``` ] .panel2-the_chunk3-auto[ ``` # A tibble: 2 × 3 pt_id time_from_initial_infusion infusion_type <chr> <dbl> <chr> 1 01 0 Initial Infusion 2 02 0 Initial Infusion ``` ] <style> .panel1-the_chunk3-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-the_chunk3-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-the_chunk3-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-the_chunk4-auto[ ``` r *end_study_events %>% head(2) ``` ] .panel2-the_chunk4-auto[ ``` # A tibble: 2 × 4 pt_id time_from_initial_infusion end_study_label end_study_name <chr> <dbl> <chr> <chr> 1 01 2 ⚠️ Other End Study Reason 2 02 0.2 ❌ Deceased ``` ] --- count: false .panel1-the_chunk4-auto[ ``` r end_study_events %>% head(2) *p3 ``` ] .panel2-the_chunk4-auto[ ``` # A tibble: 2 × 4 pt_id time_from_initial_infusion end_study_label end_study_name <chr> <dbl> <chr> <chr> 1 01 2 ⚠️ Other End Study Reason 2 02 0.2 ❌ Deceased ``` <!-- --> ] --- count: false .panel1-the_chunk4-auto[ ``` r end_study_events %>% head(2) p3 + * geom_swim_label( * data = end_study_events, * aes(x = time_from_initial_infusion, * y = pt_id, * label_vals = end_study_label, * label_names = end_study_name), * label.size = NA, fill = NA, size = 5 * ) ``` ] .panel2-the_chunk4-auto[ ``` # A tibble: 2 × 4 pt_id time_from_initial_infusion end_study_label end_study_name <chr> <dbl> <chr> <chr> 1 01 2 ⚠️ Other End Study Reason 2 02 0.2 ❌ Deceased ``` <!-- --> ] --- count: false .panel1-the_chunk4-auto[ ``` r end_study_events %>% head(2) p3 + geom_swim_label( data = end_study_events, aes(x = time_from_initial_infusion, y = pt_id, label_vals = end_study_label, label_names = end_study_name), label.size = NA, fill = NA, size = 5 ) + * scale_color_brewer(name = "Markers", palette = "Set2") ``` ] .panel2-the_chunk4-auto[ ``` # A tibble: 2 × 4 pt_id time_from_initial_infusion end_study_label end_study_name <chr> <dbl> <chr> <chr> 1 01 2 ⚠️ Other End Study Reason 2 02 0.2 ❌ Deceased ``` 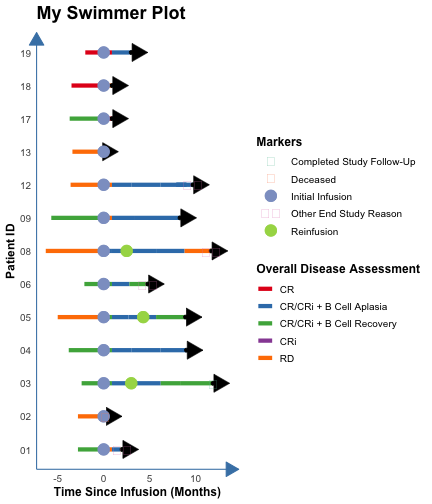<!-- --> ] <style> .panel1-the_chunk4-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-the_chunk4-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-the_chunk4-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> <style type="text/css"> .remark-code{line-height: 1.5; font-size: 80%} @media print { .has-continuation { display: block; } } code.r.hljs.remark-code{ position: relative; overflow-x: hidden; } code.r.hljs.remark-code:hover{ overflow-x:visible; width: 500px; border-style: solid; } </style>