class: center, middle, inverse, title-slide # Visualizing covariance, variance, standard deviation, correlation ### <br><br>Gina Reynolds ### <br> --- This book looks some basic statistics: - covariance - variance - standard deviation - correlation coefficient --- We'll look at the *population* statistics first equation over all: --- class: inverse, center, middle # Covariance --- class: center, middle ### Covariance is a measure of the joint variability of two random variables. --- class: inverse, center, middle .huge[ $$ cov(x,y) = \frac{\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)}{n} $$ ] --- count: false .panel1-cov_steps-auto[ ] .panel2-cov_steps-auto[ <!-- --> ] --- count: false .panel1-cov_steps-auto[ ## `$$\mu_x$$` ] .panel2-cov_steps-auto[ <!-- --> ] --- count: false .panel1-cov_steps-auto[ ## `$$\mu_y$$` ] .panel2-cov_steps-auto[ <!-- --> ] --- count: false .panel1-cov_steps-auto[ ## `$$x_i-\mu_x$$` ] .panel2-cov_steps-auto[ <!-- --> ] --- count: false .panel1-cov_steps-auto[ ## `$$y_i-\mu_y$$` ] .panel2-cov_steps-auto[ 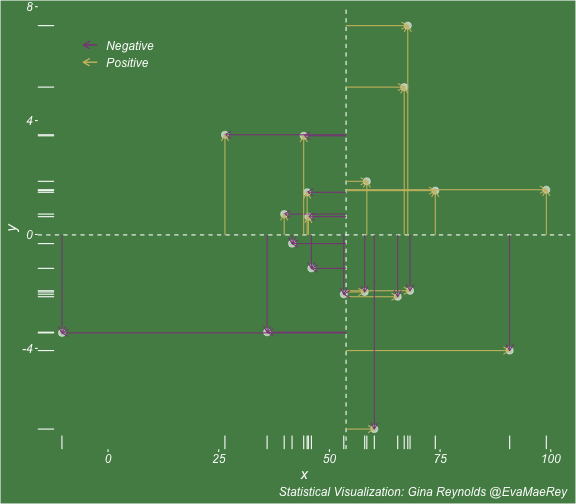<!-- --> ] --- count: false .panel1-cov_steps-auto[ ## `$$\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)$$` ] .panel2-cov_steps-auto[ 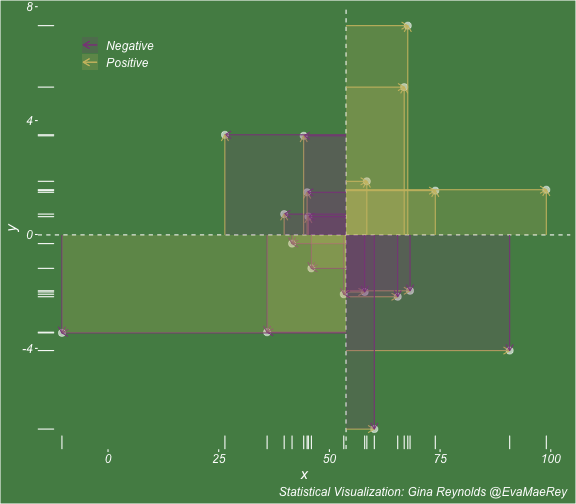<!-- --> ] --- count: false .panel1-cov_steps-auto[ ## `$$\frac{\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)}{n}$$` ] .panel2-cov_steps-auto[ 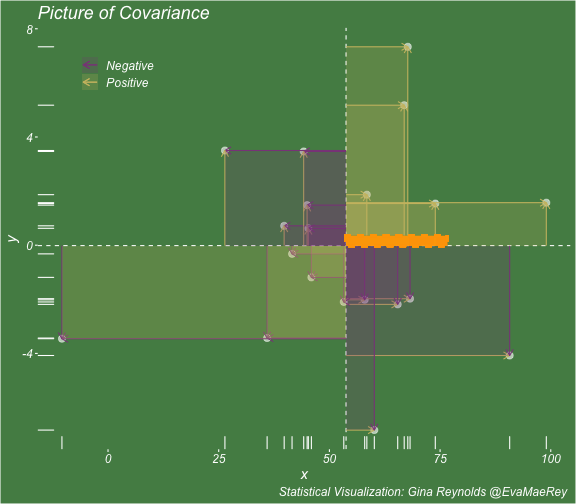<!-- --> ] <style> .panel1-cov_steps-auto { color: black; width: 43.5555555555556%; float: left; padding-left: 1%; font-size: 80% } .panel2-cov_steps-auto { color: black; width: 54.4444444444444%; float: left; padding-left: 1%; font-size: 80% } .panel3-cov_steps-auto { color: black; width: NA%; float: left; padding-left: 1%; font-size: 80% } </style> --- class: inverse, middle, center # Variance and Standard Deviation --- class: inverse, center, middle .huge[ $$ var(x) = \frac{\sum_{i=1}^n (x_i-\mu_x)^2}{n} $$ ] --- class: inverse, center, middle .huge[ $$ \sigma^2 = \frac{\sum_{i=1}^n (x_i-\mu_x)^2}{n} $$ ] --- count: false .panel1-variance_steps-user[ ] .panel2-variance_steps-user[ 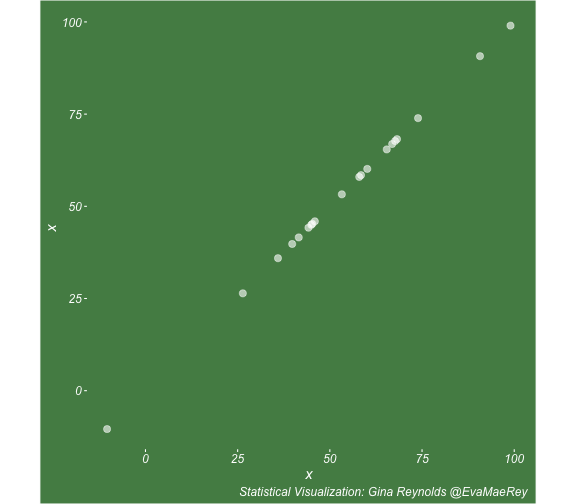<!-- --> ] --- count: false .panel1-variance_steps-user[ ## `$$\mu_x$$` ] .panel2-variance_steps-user[ 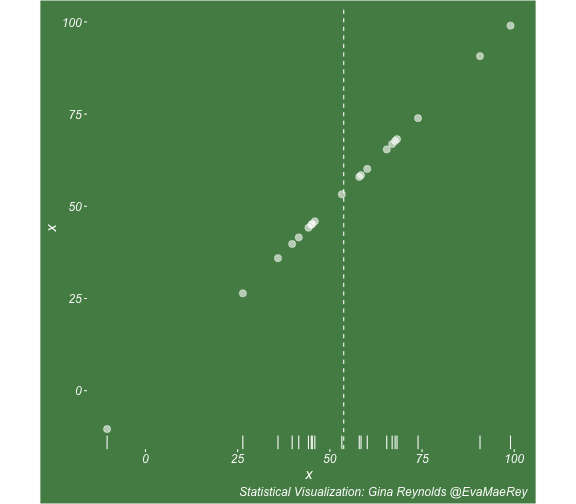<!-- --> ] --- count: false .panel1-variance_steps-user[ ## `$$\mu_x$$` ] .panel2-variance_steps-user[ 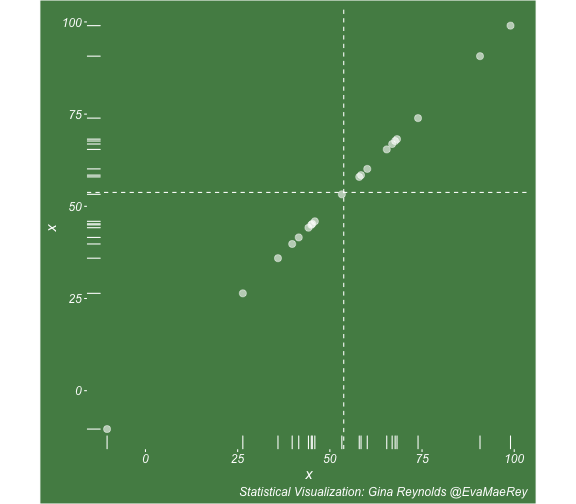<!-- --> ] --- count: false .panel1-variance_steps-user[ ## `$$x_i-\mu_x$$` ] .panel2-variance_steps-user[ 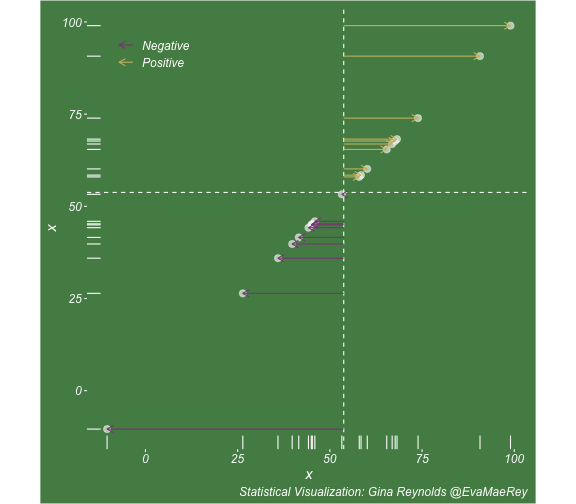<!-- --> ] --- count: false .panel1-variance_steps-user[ ## `$$x_i-\mu_x$$` ] .panel2-variance_steps-user[ 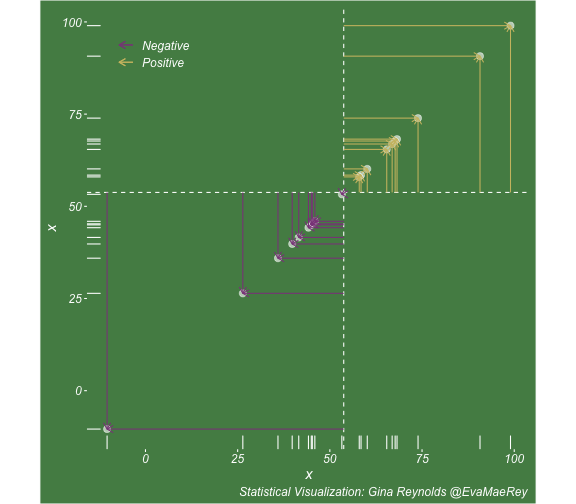<!-- --> ] --- count: false .panel1-variance_steps-user[ ## `$$\sum_{i=1}^n (x_i-\mu_x)^2$$` ] .panel2-variance_steps-user[ <!-- --> ] --- count: false .panel1-variance_steps-user[ ## `$$\frac{\sum_{i=1}^n (x_i-\mu_x)^2}{n}$$` ] .panel2-variance_steps-user[ <!-- --> ] <style> .panel1-variance_steps-user { color: black; width: 38.6060606060606%; float: left; padding-left: 1%; font-size: 80% } .panel2-variance_steps-user { color: black; width: 59.3939393939394%; float: left; padding-left: 1%; font-size: 80% } .panel3-variance_steps-user { color: black; width: NA%; float: left; padding-left: 1%; font-size: 80% } </style> --- class: inverse, center, middle .huge[ $$ \sigma^2 = \frac{\sum_{i=1}^n (x_i-\mu_x)^2}{n} $$ ] --- class: inverse, center, middle .huge[ $$ \sigma = \sqrt\frac{\sum_{i=1}^n (x_i-\mu_x)^2}{n} $$ ] --- count: false .panel1-sd_steps-user[ ## `$$\frac{\sum_{i=1}^n (x_i-\mu_x)^2}{n}$$` ] .panel2-sd_steps-user[ <!-- --> ] --- count: false .panel1-sd_steps-user[ ## `$$\sqrt\frac{\sum_{i=1}^n (x_i-\mu_x)^2}{n}$$` ] .panel2-sd_steps-user[ <!-- --> ] <style> .panel1-sd_steps-user { color: black; width: 38.6060606060606%; float: left; padding-left: 1%; font-size: 80% } .panel2-sd_steps-user { color: black; width: 59.3939393939394%; float: left; padding-left: 1%; font-size: 80% } .panel3-sd_steps-user { color: black; width: NA%; float: left; padding-left: 1%; font-size: 80% } </style> --- 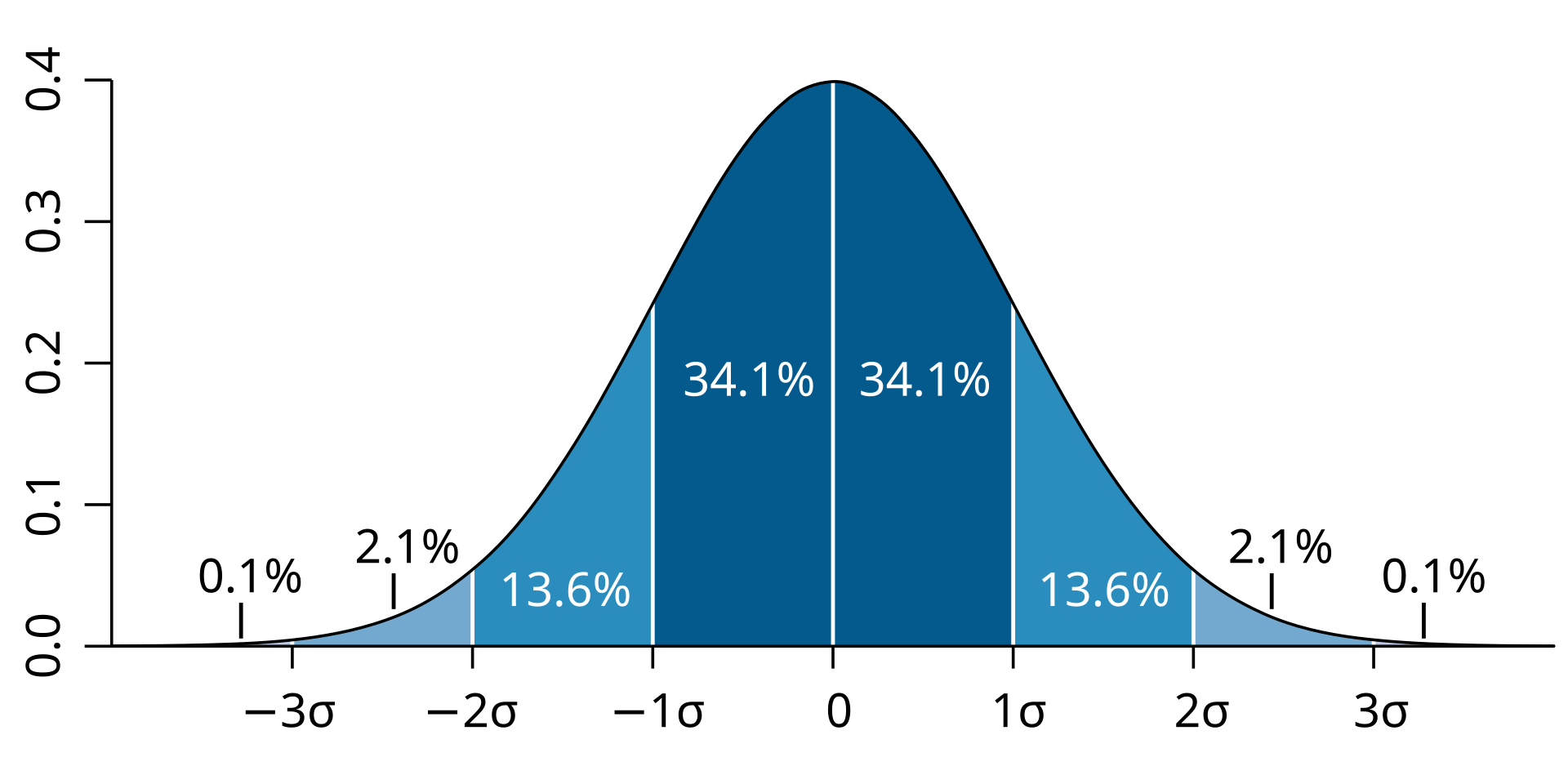 Creative commons, Wikipedia, M. W. Toews - Own work, based (in concept) on figure by Jeremy Kemp, on 2005-02-09 --- class: inverse, middle, center # correlation coefficient --- class: inverse, center, middle .huge[ $$ cor(x,y) = \frac{\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)}{n *\sigma_x \sigma_y} $$ ] --- count: false .panel1-cor_steps-user[ ## `$$\frac{\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)}{n}$$` ] .panel2-cor_steps-user[ 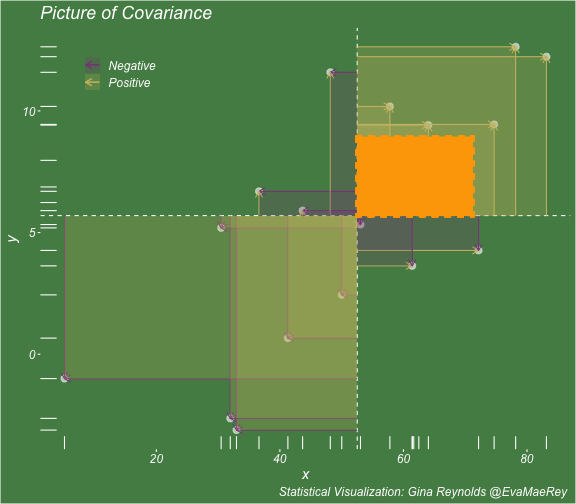<!-- --> ] --- count: false .panel1-cor_steps-user[ ## `$$\frac{\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)}{n*\sigma_x}$$` ] .panel2-cor_steps-user[ 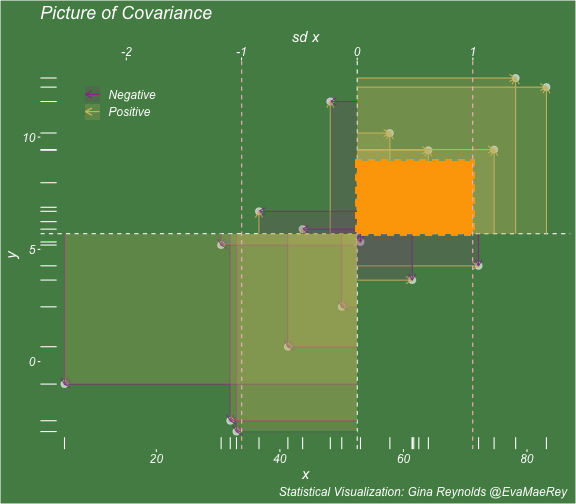<!-- --> ] --- count: false .panel1-cor_steps-user[ ## `$$\frac{\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)}{n*\sigma_x\sigma_y}$$` ] .panel2-cor_steps-user[ 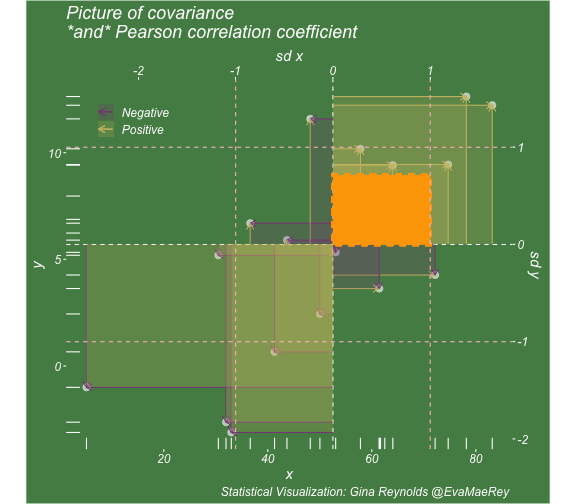<!-- --> ] --- count: false .panel1-cor_steps-user[ ## `$$\frac{\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)}{n*\sigma_x\sigma_y}$$` ] .panel2-cor_steps-user[ <!-- --> ] <style> .panel1-cor_steps-user { color: black; width: 43.5555555555556%; float: left; padding-left: 1%; font-size: 80% } .panel2-cor_steps-user { color: black; width: 54.4444444444444%; float: left; padding-left: 1%; font-size: 80% } .panel3-cor_steps-user { color: black; width: NA%; float: left; padding-left: 1%; font-size: 80% } </style> --- A negative correlation --- count: false .panel1-cor_steps_neg-user[ ## `$$\frac{\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)}{n}$$` ] .panel2-cor_steps_neg-user[ <!-- --> ] --- count: false .panel1-cor_steps_neg-user[ ## `$$\frac{\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)}{n*\sigma_x}$$` ] .panel2-cor_steps_neg-user[ <!-- --> ] --- count: false .panel1-cor_steps_neg-user[ ## `$$\frac{\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)}{n*\sigma_x\sigma_y}$$` ] .panel2-cor_steps_neg-user[ <!-- --> ] --- count: false .panel1-cor_steps_neg-user[ ## `$$\frac{\sum_{i=1}^n (x_i-\mu_x)(y_i-\mu_y)}{n*\sigma_x\sigma_y}$$` ] .panel2-cor_steps_neg-user[ <!-- --> ] <style> .panel1-cor_steps_neg-user { color: black; width: 43.5555555555556%; float: left; padding-left: 1%; font-size: 80% } .panel2-cor_steps_neg-user { color: black; width: 54.4444444444444%; float: left; padding-left: 1%; font-size: 80% } .panel3-cor_steps_neg-user { color: black; width: NA%; float: left; padding-left: 1%; font-size: 80% } </style> --- # Bessel correction: n - 1 *Sample* statistics (Not *population* statistics) --- class: inverse, center, middle .huge[ $$ cov(x,y) = \frac{\sum_{i=1}^n (x_i-\overline{x})(y_i-\overline{y})}{n - 1} $$ ] --- class: inverse, center, middle .huge[ $$ s^2 = \frac{\sum_{i=1}^n (x_i-\overline{x})^2}{n - 1} $$ ] --- class: inverse, center, middle .huge[ $$ s = \sqrt\frac{\sum_{i=1}^n (x_i-\overline{x})^2}{n - 1} $$ ] --- # DRY: Bonus -- ## "Don't repeat yourself" -- ## Writing and using functions --- ```r # this is a temporary fix - a mysetseed <- set.seed ``` --- count: false .panel1-cov_steps1-auto[ ```r *mysetseed(199402) ``` ] .panel2-cov_steps1-auto[ ] --- count: false .panel1-cov_steps1-auto[ ```r mysetseed(199402) *create_x_y(relationship = .5) ``` ] .panel2-cov_steps1-auto[ ``` # A tibble: 20 x 15 x y mean_x mean_y area mean_area quasi_mean_area sd_x_sample <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 68.2 32.1 53.8 26.9 76.1 269. 4.00 23.4 2 45.2 23.2 53.8 26.9 30.8 269. 1.62 23.4 3 26.4 16.7 53.8 26.9 278. 269. 14.6 23.4 4 90.7 41.3 53.8 26.9 533. 269. 28.0 23.4 5 58.0 27.0 53.8 26.9 0.420 269. 0.0221 23.4 6 41.6 20.5 53.8 26.9 78.2 269. 4.11 23.4 7 67.7 41.2 53.8 26.9 200. 269. 10.5 23.4 8 -10.4 -8.66 53.8 26.9 2280. 269. 120. 23.4 9 65.4 30.5 53.8 26.9 42.7 269. 2.25 23.4 10 53.3 24.5 53.8 26.9 1.20 269. 0.0634 23.4 11 58.5 31.1 53.8 26.9 19.8 269. 1.04 23.4 12 45.9 21.8 53.8 26.9 39.9 269. 2.10 23.4 13 73.9 38.5 53.8 26.9 234. 269. 12.3 23.4 14 60.1 23.2 53.8 26.9 -23.1 269. -1.22 23.4 15 99.0 51.1 53.8 26.9 1094. 269. 57.6 23.4 16 44.2 25.6 53.8 26.9 12.5 269. 0.656 23.4 17 45.0 24.0 53.8 26.9 25.5 269. 1.34 23.4 18 35.9 14.5 53.8 26.9 220. 269. 11.6 23.4 19 39.8 20.6 53.8 26.9 87.5 269. 4.60 23.4 20 66.9 38.6 53.8 26.9 154. 269. 8.12 23.4 # … with 7 more variables: sd_x <dbl>, sd_y_sample <dbl>, sd_y <dbl>, # some_x <dbl>, some_x_sample <dbl>, some_y <dbl>, some_y_sample <dbl> ``` ] --- count: false .panel1-cov_steps1-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% * data_create_scatterplot() ``` ] .panel2-cov_steps1-auto[ <!-- --> ] --- count: false .panel1-cov_steps1-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% * plot_draw_mean_x() ``` ] .panel2-cov_steps1-auto[ <!-- --> ] --- count: false .panel1-cov_steps1-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% plot_draw_mean_x() %>% * plot_draw_mean_y() ``` ] .panel2-cov_steps1-auto[ 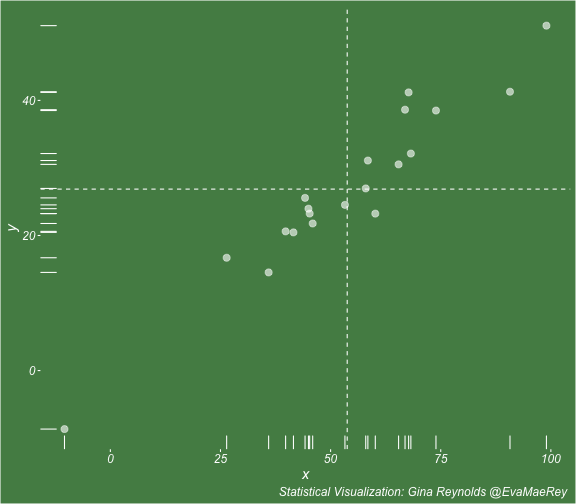<!-- --> ] --- count: false .panel1-cov_steps1-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% plot_draw_mean_x() %>% plot_draw_mean_y() %>% * plot_draw_differences_x() ``` ] .panel2-cov_steps1-auto[ 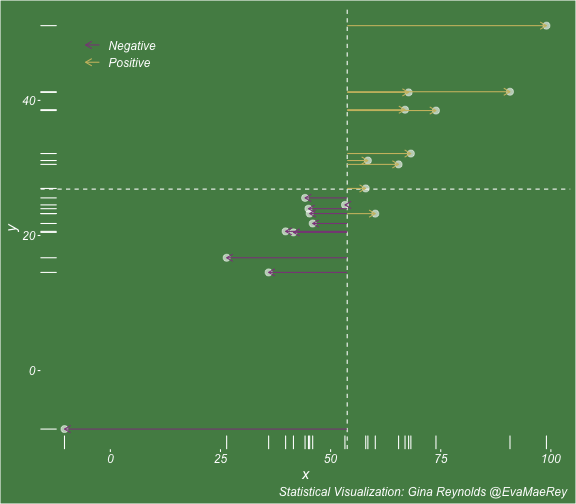<!-- --> ] --- count: false .panel1-cov_steps1-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% plot_draw_mean_x() %>% plot_draw_mean_y() %>% plot_draw_differences_x() %>% * plot_draw_differences_y() ``` ] .panel2-cov_steps1-auto[ 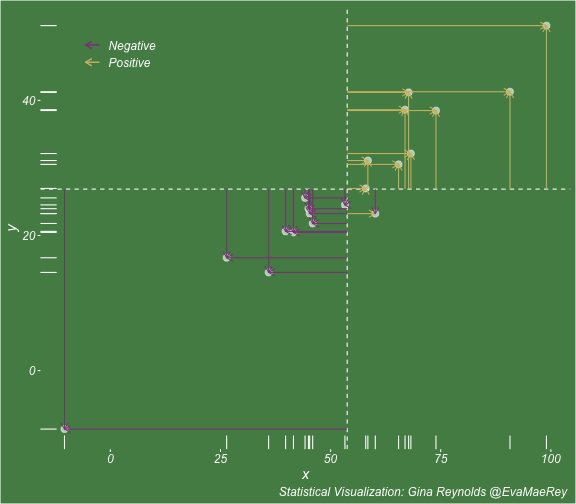<!-- --> ] --- count: false .panel1-cov_steps1-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% plot_draw_mean_x() %>% plot_draw_mean_y() %>% plot_draw_differences_x() %>% plot_draw_differences_y() %>% * plot_multiply_differences() ``` ] .panel2-cov_steps1-auto[ 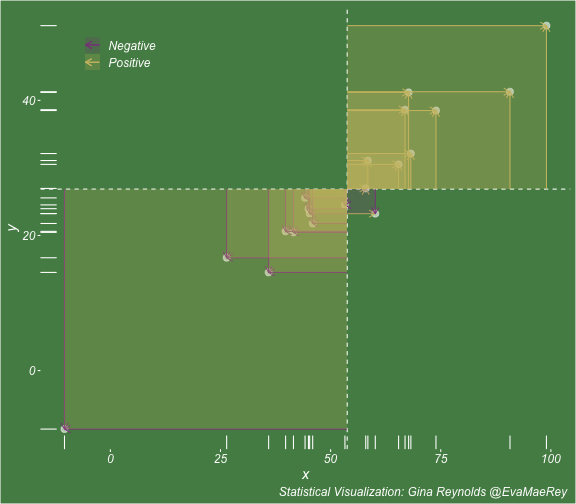<!-- --> ] --- count: false .panel1-cov_steps1-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% plot_draw_mean_x() %>% plot_draw_mean_y() %>% plot_draw_differences_x() %>% plot_draw_differences_y() %>% plot_multiply_differences() %>% * plot_take_average_rectangle() ``` ] .panel2-cov_steps1-auto[ <!-- --> ] <style> .panel1-cov_steps1-auto { color: black; width: 38.6060606060606%; float: left; padding-left: 1%; font-size: 80% } .panel2-cov_steps1-auto { color: black; width: 59.3939393939394%; float: left; padding-left: 1%; font-size: 80% } .panel3-cov_steps1-auto { color: black; width: NA%; float: left; padding-left: 1%; font-size: 80% } </style> --- count: false .panel1-cov_steps2-auto[ ```r *mysetseed(199402) ``` ] .panel2-cov_steps2-auto[ ``` function (seed, kind = NULL, normal.kind = NULL, sample.kind = NULL) { kinds <- c("Wichmann-Hill", "Marsaglia-Multicarry", "Super-Duper", "Mersenne-Twister", "Knuth-TAOCP", "user-supplied", "Knuth-TAOCP-2002", "L'Ecuyer-CMRG", "default") n.kinds <- c("Buggy Kinderman-Ramage", "Ahrens-Dieter", "Box-Muller", "user-supplied", "Inversion", "Kinderman-Ramage", "default") s.kinds <- c("Rounding", "Rejection", "default") if (length(kind)) { if (!is.character(kind) || length(kind) > 1L) stop("'kind' must be a character string of length 1 (RNG to be used).") if (is.na(i.knd <- pmatch(kind, kinds) - 1L)) stop(gettextf("'%s' is not a valid abbreviation of an RNG", kind), domain = NA) if (i.knd == length(kinds) - 1L) i.knd <- -1L } else i.knd <- NULL if (!is.null(normal.kind)) { if (!is.character(normal.kind) || length(normal.kind) != 1L) stop("'normal.kind' must be a character string of length 1") normal.kind <- pmatch(normal.kind, n.kinds) - 1L if (is.na(normal.kind)) stop(gettextf("'%s' is not a valid choice", normal.kind), domain = NA) if (normal.kind == 0L) stop("buggy version of Kinderman-Ramage generator is not allowed", domain = NA) if (normal.kind == length(n.kinds) - 1L) normal.kind <- -1L } if (!is.null(sample.kind)) { if (!is.character(sample.kind) || length(sample.kind) != 1L) stop("'sample.kind' must be a character string of length 1") sample.kind <- pmatch(sample.kind, s.kinds) - 1L if (is.na(sample.kind)) stop(gettextf("'%s' is not a valid choice", sample.kind), domain = NA) if (sample.kind == 0L) warning("non-uniform 'Rounding' sampler used", domain = NA) if (sample.kind == length(s.kinds) - 1L) sample.kind <- -1L } .Internal(set.seed(seed, i.knd, normal.kind, sample.kind)) } <bytecode: 0x7fda69ccf8b8> <environment: namespace:base> ``` ] .panel3-cov_steps2-auto[ ] --- count: false .panel1-cov_steps2-auto[ ```r mysetseed(199402) *create_x_y(relationship = .5) ``` ] .panel2-cov_steps2-auto[ ``` function(num = 20, spread_x = 20, relationship = .1, noise = 3 ){ tibble(x = rnorm(num, sd = spread_x) + 50 ) %>% mutate(y = relationship * x + rnorm(num, sd = noise)) %>% mutate(mean_x = mean(x)) %>% mutate(mean_y = mean(y)) %>% mutate(area = (x - mean_x)*(y - mean_y)) %>% mutate(mean_area = mean(area)) %>% mutate(quasi_mean_area = area/(n() - 1)) %>% mutate(sd_x_sample = sd(x)) %>% mutate(sd_x = sd_pop(x)) %>% mutate(sd_y_sample = sd(y)) %>% mutate(sd_y = sd_pop(y)) %>% mutate(some_x = sd_x) %>% mutate(some_x_sample = sd_x_sample) %>% mutate(some_y = mean_area/some_x) %>% mutate(some_y_sample = sd_y_sample) } <bytecode: 0x7fda6c9d67c0> ``` ] .panel3-cov_steps2-auto[ ``` # A tibble: 20 x 15 x y mean_x mean_y area mean_area quasi_mean_area sd_x_sample <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 68.2 32.1 53.8 26.9 76.1 269. 4.00 23.4 2 45.2 23.2 53.8 26.9 30.8 269. 1.62 23.4 3 26.4 16.7 53.8 26.9 278. 269. 14.6 23.4 4 90.7 41.3 53.8 26.9 533. 269. 28.0 23.4 5 58.0 27.0 53.8 26.9 0.420 269. 0.0221 23.4 6 41.6 20.5 53.8 26.9 78.2 269. 4.11 23.4 7 67.7 41.2 53.8 26.9 200. 269. 10.5 23.4 8 -10.4 -8.66 53.8 26.9 2280. 269. 120. 23.4 9 65.4 30.5 53.8 26.9 42.7 269. 2.25 23.4 10 53.3 24.5 53.8 26.9 1.20 269. 0.0634 23.4 11 58.5 31.1 53.8 26.9 19.8 269. 1.04 23.4 12 45.9 21.8 53.8 26.9 39.9 269. 2.10 23.4 13 73.9 38.5 53.8 26.9 234. 269. 12.3 23.4 14 60.1 23.2 53.8 26.9 -23.1 269. -1.22 23.4 15 99.0 51.1 53.8 26.9 1094. 269. 57.6 23.4 16 44.2 25.6 53.8 26.9 12.5 269. 0.656 23.4 17 45.0 24.0 53.8 26.9 25.5 269. 1.34 23.4 18 35.9 14.5 53.8 26.9 220. 269. 11.6 23.4 19 39.8 20.6 53.8 26.9 87.5 269. 4.60 23.4 20 66.9 38.6 53.8 26.9 154. 269. 8.12 23.4 # … with 7 more variables: sd_x <dbl>, sd_y_sample <dbl>, sd_y <dbl>, # some_x <dbl>, some_x_sample <dbl>, some_y <dbl>, some_y_sample <dbl> ``` ] --- count: false .panel1-cov_steps2-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% * data_create_scatterplot() ``` ] .panel2-cov_steps2-auto[ ``` function(data, background_color = "palegreen4"){ data %>% ggplot() + theme(legend.position = c(.15, .9)) + aes(x = x) + aes(y = y) + theme(rect = element_rect(fill = background_color)) + theme(text = element_text(color = "white", face = "italic", size = 15)) + theme(panel.background = element_rect(fill = background_color)) + theme(legend.key = element_blank()) + #element_rect(fill = background_color)) + theme(legend.title = element_blank()) + theme(axis.text = element_text(color = "white")) + theme(axis.ticks = element_line(color = "white")) + labs(title = NULL) + theme(panel.grid = element_blank()) + geom_point(size = 3, pch = 21, col = "white", fill = "white", lwd = 3, alpha = .6) + labs(caption = "Statistical Visualization: Gina Reynolds @EvaMaeRey ") } <bytecode: 0x7fda6d7f4c98> ``` ] .panel3-cov_steps2-auto[ <!-- --> ] --- count: false .panel1-cov_steps2-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% * plot_draw_mean_x() ``` ] .panel2-cov_steps2-auto[ ``` function(plot){ plot + # 1. mean of x geom_rug(aes(y = NULL), col = "white") + geom_vline(aes(xintercept = mean(x)), lty = "dashed", col = "white") } <bytecode: 0x7fda6fb08208> ``` ] .panel3-cov_steps2-auto[ <!-- --> ] --- count: false .panel1-cov_steps2-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% plot_draw_mean_x() %>% * plot_draw_mean_y() ``` ] .panel2-cov_steps2-auto[ ``` function(plot){ plot + # 2. mean of y geom_rug(col = "white", aes(x = NULL)) + geom_hline(aes(yintercept = mean(y)), lty = "dashed", col = "white") } <bytecode: 0x7fda6c673368> ``` ] .panel3-cov_steps2-auto[ <!-- --> ] --- count: false .panel1-cov_steps2-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% plot_draw_mean_x() %>% plot_draw_mean_y() %>% * plot_draw_differences_x() ``` ] .panel2-cov_steps2-auto[ ``` function(plot){ plot + # difference xi mean x scale_color_manual(breaks = c(FALSE, TRUE), label = c("Negative", "Positive"), values = c("orchid4", "lightgoldenrod3") ) + geom_segment(aes(col = x > mean(x), xend = mean(x), yend = y), arrow = arrow(ends = "first", length = unit(0.1, "inches"))) + labs(col = "") } <bytecode: 0x7fda6bcca1c8> ``` ] .panel3-cov_steps2-auto[ <!-- --> ] --- count: false .panel1-cov_steps2-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% plot_draw_mean_x() %>% plot_draw_mean_y() %>% plot_draw_differences_x() %>% * plot_draw_differences_y() ``` ] .panel2-cov_steps2-auto[ ``` function(plot){ plot + # difference yi mean y geom_segment(aes(col = y > mean(y), xend = x, yend = mean(y)), arrow = arrow(ends = "first", length = unit(0.1, "inches"))) } <bytecode: 0x7fda6d6932e0> ``` ] .panel3-cov_steps2-auto[ <!-- --> ] --- count: false .panel1-cov_steps2-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% plot_draw_mean_x() %>% plot_draw_mean_y() %>% plot_draw_differences_x() %>% plot_draw_differences_y() %>% * plot_multiply_differences() ``` ] .panel2-cov_steps2-auto[ ``` function(plot){ plot + # multiply differences aes(fill = area > 0) + geom_rect(aes(xmin = mean(x), ymin = mean(y), ymax = y, xmax = x), alpha = .2 ) + labs(fill = "") + scale_fill_manual(breaks = c(FALSE, TRUE), label = c("Negative", "Positive"), values = c("orchid4", "lightgoldenrod3")) } <bytecode: 0x7fda6e4e5698> ``` ] .panel3-cov_steps2-auto[ <!-- --> ] --- count: false .panel1-cov_steps2-auto[ ```r mysetseed(199402) create_x_y(relationship = .5) %>% data_create_scatterplot() %>% plot_draw_mean_x() %>% plot_draw_mean_y() %>% plot_draw_differences_x() %>% plot_draw_differences_y() %>% plot_multiply_differences() %>% * plot_take_average_rectangle() ``` ] .panel2-cov_steps2-auto[ ``` function(plot, title = "Picture of Covariance"){ plot + # Average areas geom_rect(aes( xmin = mean(x), ymin = mean(y), ymax = mean(some_y) + mean(y), xmax = mean(some_x) + mean(x) ), color = "orange", linetype = "dotted", fill = "orange", lwd = 1.5, alpha = .2) + #BREAK labs(title = title) } ``` ] .panel3-cov_steps2-auto[ <!-- --> ] <style> .panel1-cov_steps2-auto { color: black; width: 27.28125%; float: left; padding-left: 1%; font-size: 80% } .panel2-cov_steps2-auto { color: black; width: 37.3854166666667%; float: left; padding-left: 1%; font-size: 80% } .panel3-cov_steps2-auto { color: black; width: 32.3333333333333%; float: left; padding-left: 1%; font-size: 80% } </style> --- --- # DRY next level: writing and using packages - [A companion guide to Jim Hester’s, 'You can make an R package in 20 minutes'](https://evamaerey.github.io/package_in_20_minutes/package_in_20_minutes) --- count: false .panel1-numeric-auto[ ```r *cars ``` ] .panel2-numeric-auto[ ``` speed dist 1 4 2 2 4 10 3 7 4 4 7 22 5 8 16 6 9 10 7 10 18 8 10 26 9 10 34 10 11 17 11 11 28 12 12 14 13 12 20 14 12 24 15 12 28 16 13 26 17 13 34 18 13 34 19 13 46 20 14 26 21 14 36 22 14 60 23 14 80 24 15 20 25 15 26 26 15 54 27 16 32 28 16 40 29 17 32 30 17 40 31 17 50 32 18 42 33 18 56 34 18 76 35 18 84 36 19 36 37 19 46 38 19 68 39 20 32 40 20 48 41 20 52 42 20 56 43 20 64 44 22 66 45 23 54 46 24 70 47 24 92 48 24 93 49 24 120 50 25 85 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% * ggplot() ``` ] .panel2-numeric-auto[ <!-- --> ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + * aes(x = speed, y = dist) ``` ] .panel2-numeric-auto[ <!-- --> ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + * geom_point() ``` ] .panel2-numeric-auto[ <!-- --> ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> *visualization ``` ] .panel2-numeric-auto[ ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation *cars$speed ``` ] .panel2-numeric-auto[ ``` [1] 4 4 7 7 8 9 10 10 10 11 11 12 12 12 12 13 13 13 13 14 14 14 14 15 15 [26] 15 16 16 17 17 17 18 18 18 18 19 19 19 20 20 20 20 20 22 23 24 24 24 24 25 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - * mean(cars$speed) ``` ] .panel2-numeric-auto[ ``` [1] -11.4 -11.4 -8.4 -8.4 -7.4 -6.4 -5.4 -5.4 -5.4 -4.4 -4.4 -3.4 [13] -3.4 -3.4 -3.4 -2.4 -2.4 -2.4 -2.4 -1.4 -1.4 -1.4 -1.4 -0.4 [25] -0.4 -0.4 0.6 0.6 1.6 1.6 1.6 2.6 2.6 2.6 2.6 3.6 [37] 3.6 3.6 4.6 4.6 4.6 4.6 4.6 6.6 7.6 8.6 8.6 8.6 [49] 8.6 9.6 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> *x_diff ``` ] .panel2-numeric-auto[ ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff *cars$dist ``` ] .panel2-numeric-auto[ ``` [1] 2 10 4 22 16 10 18 26 34 17 28 14 20 24 28 26 34 34 46 [20] 26 36 60 80 20 26 54 32 40 32 40 50 42 56 76 84 36 46 68 [39] 32 48 52 56 64 66 54 70 92 93 120 85 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff cars$dist - * mean(cars$dist) ``` ] .panel2-numeric-auto[ ``` [1] -40.98 -32.98 -38.98 -20.98 -26.98 -32.98 -24.98 -16.98 -8.98 -25.98 [11] -14.98 -28.98 -22.98 -18.98 -14.98 -16.98 -8.98 -8.98 3.02 -16.98 [21] -6.98 17.02 37.02 -22.98 -16.98 11.02 -10.98 -2.98 -10.98 -2.98 [31] 7.02 -0.98 13.02 33.02 41.02 -6.98 3.02 25.02 -10.98 5.02 [41] 9.02 13.02 21.02 23.02 11.02 27.02 49.02 50.02 77.02 42.02 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff cars$dist - mean(cars$dist) -> *y_diff ``` ] .panel2-numeric-auto[ ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff cars$dist - mean(cars$dist) -> y_diff *x_diff ``` ] .panel2-numeric-auto[ ``` [1] -11.4 -11.4 -8.4 -8.4 -7.4 -6.4 -5.4 -5.4 -5.4 -4.4 -4.4 -3.4 [13] -3.4 -3.4 -3.4 -2.4 -2.4 -2.4 -2.4 -1.4 -1.4 -1.4 -1.4 -0.4 [25] -0.4 -0.4 0.6 0.6 1.6 1.6 1.6 2.6 2.6 2.6 2.6 3.6 [37] 3.6 3.6 4.6 4.6 4.6 4.6 4.6 6.6 7.6 8.6 8.6 8.6 [49] 8.6 9.6 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff cars$dist - mean(cars$dist) -> y_diff x_diff %>% * `*`(y_diff) ``` ] .panel2-numeric-auto[ ``` [1] 467.172 375.972 327.432 176.232 199.652 211.072 134.892 91.692 48.492 [10] 114.312 65.912 98.532 78.132 64.532 50.932 40.752 21.552 21.552 [19] -7.248 23.772 9.772 -23.828 -51.828 9.192 6.792 -4.408 -6.588 [28] -1.788 -17.568 -4.768 11.232 -2.548 33.852 85.852 106.652 -25.128 [37] 10.872 90.072 -50.508 23.092 41.492 59.892 96.692 151.932 83.752 [46] 232.372 421.572 430.172 662.372 403.392 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff cars$dist - mean(cars$dist) -> y_diff x_diff %>% `*`(y_diff) %>% * sum() ``` ] .panel2-numeric-auto[ ``` [1] 5387.4 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff cars$dist - mean(cars$dist) -> y_diff x_diff %>% `*`(y_diff) %>% sum() %>% * `/`(nrow(cars) - 1) ``` ] .panel2-numeric-auto[ ``` [1] 109.9469 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff cars$dist - mean(cars$dist) -> y_diff x_diff %>% `*`(y_diff) %>% sum() %>% `/`(nrow(cars) - 1) # or use the function *library(magrittr) ``` ] .panel2-numeric-auto[ ``` [1] 109.9469 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff cars$dist - mean(cars$dist) -> y_diff x_diff %>% `*`(y_diff) %>% sum() %>% `/`(nrow(cars) - 1) # or use the function library(magrittr) *cars ``` ] .panel2-numeric-auto[ ``` [1] 109.9469 ``` ``` speed dist 1 4 2 2 4 10 3 7 4 4 7 22 5 8 16 6 9 10 7 10 18 8 10 26 9 10 34 10 11 17 11 11 28 12 12 14 13 12 20 14 12 24 15 12 28 16 13 26 17 13 34 18 13 34 19 13 46 20 14 26 21 14 36 22 14 60 23 14 80 24 15 20 25 15 26 26 15 54 27 16 32 28 16 40 29 17 32 30 17 40 31 17 50 32 18 42 33 18 56 34 18 76 35 18 84 36 19 36 37 19 46 38 19 68 39 20 32 40 20 48 41 20 52 42 20 56 43 20 64 44 22 66 45 23 54 46 24 70 47 24 92 48 24 93 49 24 120 50 25 85 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff cars$dist - mean(cars$dist) -> y_diff x_diff %>% `*`(y_diff) %>% sum() %>% `/`(nrow(cars) - 1) # or use the function library(magrittr) cars %$% * cov(dist, speed) ``` ] .panel2-numeric-auto[ ``` [1] 109.9469 ``` ``` [1] 109.9469 ``` ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff cars$dist - mean(cars$dist) -> y_diff x_diff %>% `*`(y_diff) %>% sum() %>% `/`(nrow(cars) - 1) # or use the function library(magrittr) cars %$% cov(dist, speed) *visualization ``` ] .panel2-numeric-auto[ ``` [1] 109.9469 ``` ``` [1] 109.9469 ``` <!-- --> ] --- count: false .panel1-numeric-auto[ ```r cars %>% ggplot() + aes(x = speed, y = dist) + geom_point() -> visualization # 'by-hand' calculation cars$speed - mean(cars$speed) -> x_diff cars$dist - mean(cars$dist) -> y_diff x_diff %>% `*`(y_diff) %>% sum() %>% `/`(nrow(cars) - 1) # or use the function library(magrittr) cars %$% cov(dist, speed) visualization + * annotate(geom = "text", * x = 7, * y = 100, * label = paste0("cov = ", * cars %$% * cov(dist, speed) %>% * round(1))) ``` ] .panel2-numeric-auto[ ``` [1] 109.9469 ``` ``` [1] 109.9469 ``` <!-- --> ] <style> .panel1-numeric-auto { color: black; width: 38.6060606060606%; float: left; padding-left: 1%; font-size: 80% } .panel2-numeric-auto { color: black; width: 59.3939393939394%; float: left; padding-left: 1%; font-size: 80% } .panel3-numeric-auto { color: black; width: NA%; float: left; padding-left: 1%; font-size: 80% } </style> --- <img src="covariance_correlation_files/figure-html/unnamed-chunk-4-1.png" width="25%" /><img src="covariance_correlation_files/figure-html/unnamed-chunk-4-2.png" width="25%" /><img src="covariance_correlation_files/figure-html/unnamed-chunk-4-3.png" width="25%" /><img src="covariance_correlation_files/figure-html/unnamed-chunk-4-4.png" width="25%" /><img src="covariance_correlation_files/figure-html/unnamed-chunk-4-5.png" width="25%" /><img src="covariance_correlation_files/figure-html/unnamed-chunk-4-6.png" width="25%" /><img src="covariance_correlation_files/figure-html/unnamed-chunk-4-7.png" width="25%" /><img src="covariance_correlation_files/figure-html/unnamed-chunk-4-8.png" width="25%" /> <style type="text/css"> .remark-code{line-height: 1.5; font-size: 70%} .huge { font-size: 250%; } @media print { .has-continuation { display: block; } } </style>