The goal of ggsample provide functions for sampling. The facet_* functions could be especially statistical education in helping visualize hypothetical scenarios (like sampling from known population) or demonstrating resampling techniques. The facet_* functions mostly borrow from the ggplot2 extension vignette (extending existing facet function). But a seed setting move has been added such that the layers in each facet are looking at the same sampling frame; i.e. geom_smooth() will be acting on the same data as geom_point() points This which didn’t seem to be the case when only using the vignette strategy as-is. The functions are:
facet_sample()facet_sample_prop()facet_bootstrap()facet_scramble()
There are also some geom experiments, which I think might be less useful.
Installation
And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("EvaMaeRey/ggsample")Examples
Below we display 9 panels with different a sample of 16 (n_facets default) observations taken.
library(ggplot2)
library(ggsample)
# facet_sample
ggplot(data = mtcars) +
aes(x = wt) +
aes(y = mpg) +
facet_sample(n_sampled = 10) +
geom_point(alpha = .5) +
geom_smooth(method = lm, se = F)
#> `geom_smooth()` using formula = 'y ~ x'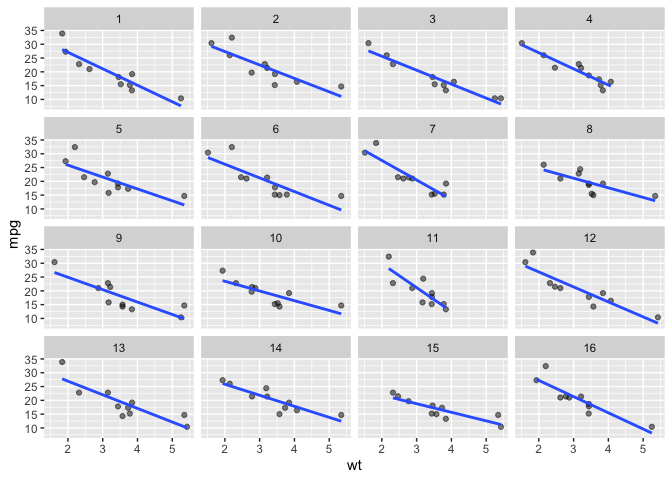
We can also sample by specifying a proportion with facet_sample_prop().
# facet_sample_prop
ggplot(data = mtcars) +
aes(x = wt) +
aes(y = mpg) +
facet_sample_prop(prop = .2,
n_facets = 9) +
geom_point(alpha = .5) +
geom_smooth(method = lm, se = F)
#> `geom_smooth()` using formula = 'y ~ x'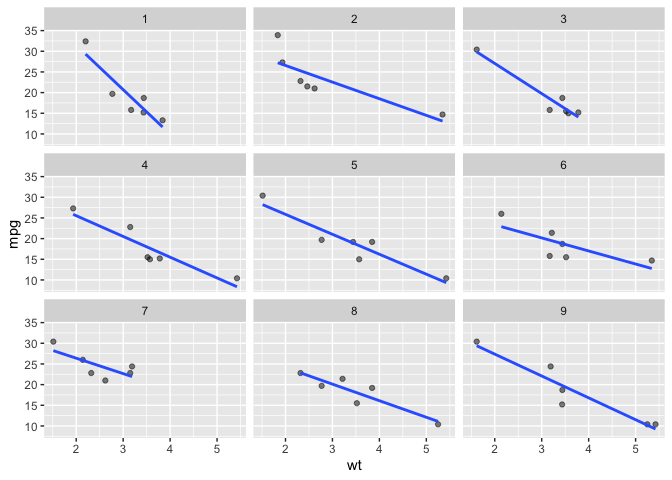
We can also bootstrap taking a sample of nrow(data) with replacement via facet_bootstrap().
# facet_bootstrap
ggplot(data = mtcars) +
aes(x = wt) +
aes(y = mpg) +
facet_bootstrap() +
geom_count(alpha = .2) +
geom_smooth(method = lm, se = F)
#> `geom_smooth()` using formula = 'y ~ x'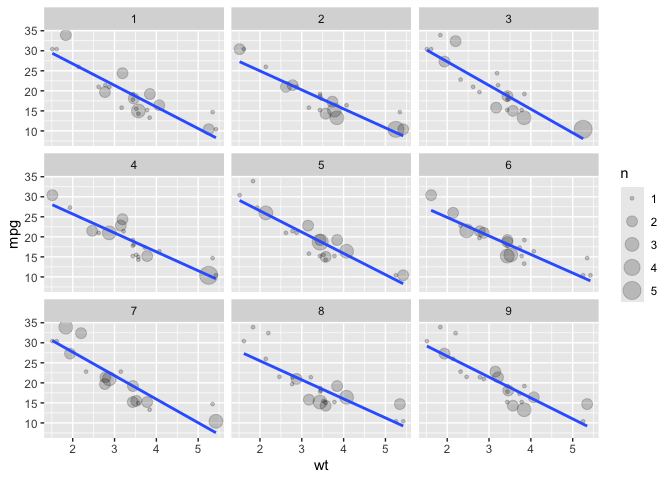
Looking at disassociated data (variables in a dataset are randomly reordered) is also possible via facet_scramble().
# facet_scramble
ggplot(data = mtcars) +
aes(x = wt) +
aes(y = mpg) +
facet_scramble(n_facets = 9) +
geom_point(alpha = .2) +
geom_smooth(method = lm, se = F)
#> `geom_smooth()` using formula = 'y ~ x'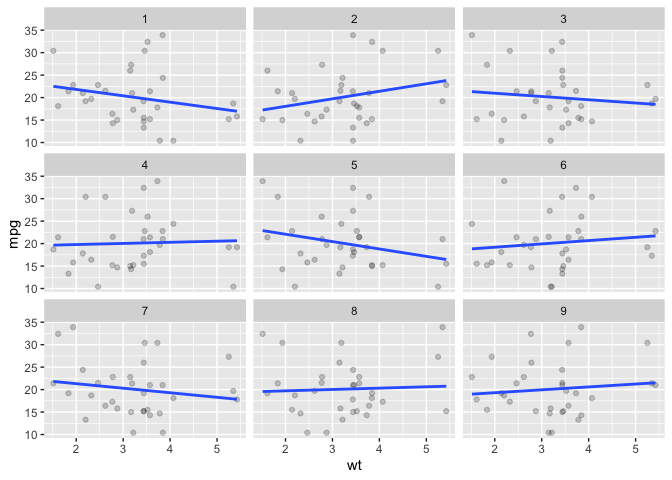
Simulating reality: single sample realization with facet_*(n_facets = 1)
You can use facet_*(n_facets = 1) to see a single realization. In a classroom setting you can re-execute code to see multiple realizations one at a time. last_plot() does not produce a fresh sample; but overriding facet_sample does work.
I wonder if some helpers might be nice like facet_sample_once() for this special case.
ggplot(data = mtcars) +
aes(x = wt) +
aes(y = mpg) +
facet_sample(n_sampled = 10,
n_facets = 1) +
geom_point(alpha = .5) +
geom_smooth(method = lm, se = F)
#> `geom_smooth()` using formula = 'y ~ x'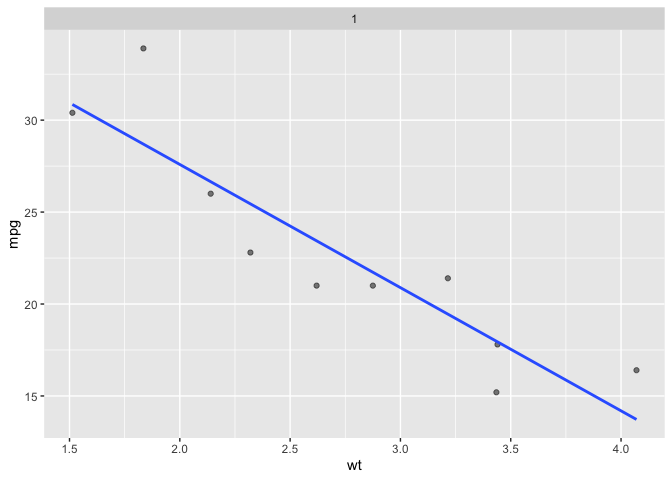
# replace facet declaration
last_plot() + facet_sample(n_sampled = 10, n_facets = 1)
#> `geom_smooth()` using formula = 'y ~ x'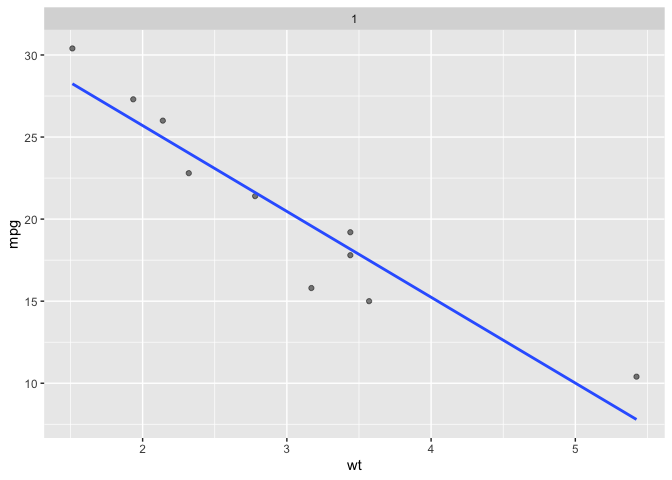
Functions internals…
Here, you’ll see a lot of cloned code from the ggplot2 extension vignette! https://github.com/tidyverse/ggplot2/blob/5f518d02af27160ab98fed736a472321d72d10d2/vignettes/extending-ggplot2.Rmd#L1028 The small contribution made here is the seed parameter which, ensures that layers are displayed based on the same sample (geom_smooth reflects the same data as geom_point) which seems to have worked.
There is a lot of repetition in what’s below and it might be good to try to consolidate some of this at some point.
compute_layout_sample <- function(data, params) {
id <- seq_len(params$n)
dims <- wrap_dims(params$n, params$nrow, params$ncol)
layout <- data.frame(PANEL = factor(id))
if (params$as.table) { layout$ROW <- as.integer((id - 1L) %/% dims[2] + 1L)
} else { layout$ROW <- as.integer(dims[1] - (id - 1L) %/% dims[2]) }
layout$COL <- as.integer((id - 1L) %% dims[2] + 1L)
layout <- layout[order(layout$PANEL), , drop = FALSE]
rownames(layout) <- NULL
# Add scale identification
layout$SCALE_X <- if (params$free$x) id else 1L
layout$SCALE_Y <- if (params$free$y) id else 1L
cbind(layout, .bootstrap = id)
}
map_data_sample <- function(data, layout, params) {
if (is.null(data) || nrow(data) == 0) {return(cbind(data, PANEL = integer(0)))}
set.seed(params$seed) # adding this
n_sampled <- params$n_sampled
new_data <- lapply(seq_len(params$n), function(i) {
cbind(data[sample(nrow(data), n_sampled), , drop = FALSE], PANEL = i)
})
do.call(rbind, new_data)
}
FacetSample <- ggplot2::ggproto(
`_class` = "FacetSample",
`_inherit` = ggplot2::FacetWrap,
compute_layout = compute_layout_sample,
map_data = map_data_sample
)
#' Title
#'
#' @param n_facets number of facets
#' @param nrow how many rows for facet grid
#' @param ncol how many columns for facet grid
#' @param scales should x and y scales be determined independently for facets
#' @param shrink not sure what shrink does
#' @param strip.position where facet label should go
#' @param n_sampled sample size
#' @param seed randomization start point
#'
#' @return a ggplot2 facet specification
#' @export
#'
#' @examples
#' # mean from samples
#' library(ggplot2)
#' ggplot(data = mtcars) +
#' aes(x = wt) +
#' aes(y = mpg) +
#' facet_sample(n_sampled = 10,
#' n_facets = 9,
#' nrow = 3) +
#' geom_point(alpha = .5) +
#' geom_smooth(method = lm, se = F)
#' labs(subtitle = "Depending on our particular random sample...")
facet_sample <- function(n_facets = 16, n_sampled = 5, nrow = NULL, ncol = NULL,
scales = "fixed", shrink = TRUE, strip.position = "top",
seed = sample(2000:3000, 1)) {
facet <- ggplot2::facet_wrap(~.bootstrap, nrow = nrow, ncol = ncol, scales = scales,
shrink = shrink, strip.position = strip.position)
facet$params$n <- n_facets
facet$params$n_sampled <- n_sampled
facet$params$seed <- seed
ggplot2::ggproto(`_class` = NULL,
`_inherit` = FacetSample,
shrink = shrink,
params = facet$params)
}try it out
library(ggplot2)
ggplot(data = mtcars) +
aes(x = wt) +
aes(y = mpg) +
facet_sample(n_sampled = 10,
n_facets = 9,
nrow = 3) +
geom_point(alpha = .5) +
geom_smooth(method = lm, se = F)
#> `geom_smooth()` using formula = 'y ~ x'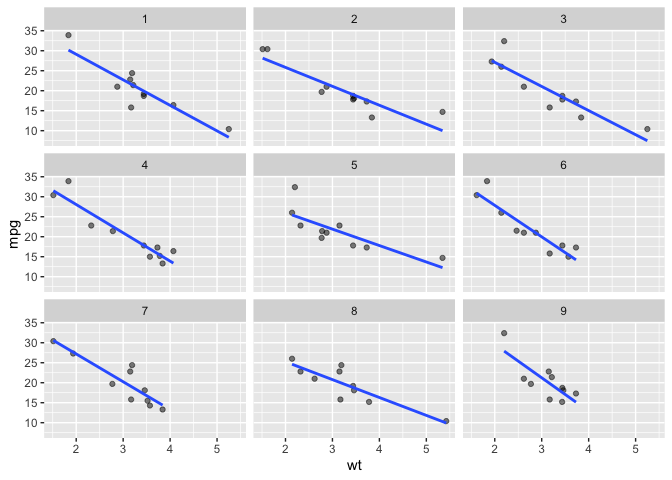
facet_sample_prop()
#' Title
#'
#' @inheritParams facet_sample
#' @param prop numeric proportion to sample, defaults to .2
#'
#' @return
#' @export
#'
#' @examples
#' set.seed(1323)
#' library(ggplot2)
#' ggplot(data = cars) +
#' aes(x = speed) +
#' aes(y = dist) +
#' geom_point(color = "olivedrab4") +
#' geom_smooth(method = lm, se = FALSE) +
#' facet_sample_prop() +
#' labs(title = "Random Sample, 20% of population")
facet_sample_prop <- function(n_facets = 9, prop = 0.2, nrow = NULL, ncol = NULL,
scales = "fixed", shrink = TRUE, strip.position = "top",
seed = sample(2000:3000, 1)) {
facet <- ggplot2::facet_wrap(~.bootstrap, nrow = nrow, ncol = ncol, scales = scales,
shrink = shrink, strip.position = strip.position)
facet$params$n <- n_facets
facet$params$prop <- prop
facet$params$seed <- seed
ggplot2::ggproto(NULL, FacetSampleProp,
shrink = shrink,
params = facet$params
)
}
map_data_sample_prop <- function(data, layout, params) {
if (is.null(data) || nrow(data) == 0) {
return(cbind(data, PANEL = integer(0)))
}
set.seed(params$seed)
n_samples <- round(nrow(data) * params$prop)
new_data <- lapply(seq_len(params$n), function(i) {
cbind(data[sample(nrow(data), n_samples), , drop = FALSE], PANEL = i)
})
do.call(rbind, new_data)
}
FacetSampleProp <- ggplot2::ggproto("FacetSampleProp", ggplot2::FacetWrap,
compute_layout = compute_layout_sample,
map_data = map_data_sample_prop
)Try it out…
# facet_sample_prop
ggplot(data = mtcars) +
aes(x = wt) +
aes(y = mpg) +
facet_sample_prop(prop = .1,
n_facets = 9,
nrow = 3) +
geom_point(alpha = .5) +
geom_smooth(method = lm, se = F)
#> `geom_smooth()` using formula = 'y ~ x'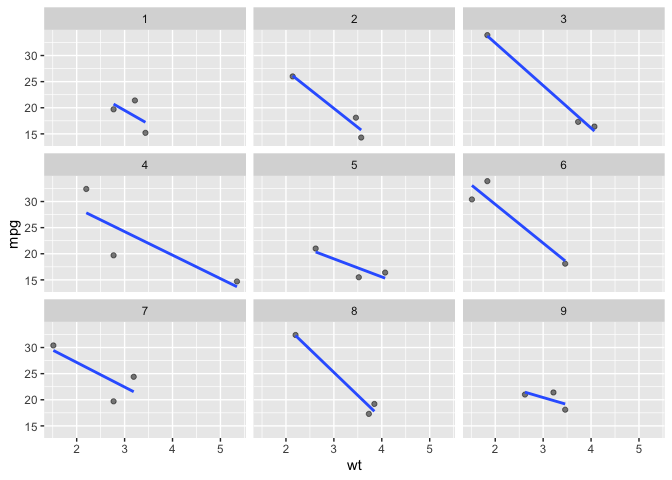
facet_bootstrap()
#' Resample with replacement
#'
#' @inheritParams facet_sample_prop
#'
#' @return
#' @export
#'
#' @examples
#' library(ggplot2)
#' set.seed(1323)
#' ggplot(data = cars) +
#' aes(x = speed) +
#' aes(y = dist) +
#' geom_count(alpha = .5) +
#' facet_bootstrap(n_facets = 1) +
#' facet_bootstrap(n_facets = 2) +
#' facet_bootstrap(n_facets = 3) +
#' ggxmean::geom_lm() +
#' ggxmean::geom_lm_formula() +
#' facet_bootstrap(n_facets = 15)
facet_bootstrap <- function(n_facets = 9, prop = 1, nrow = NULL, ncol = NULL,
scales = "fixed", shrink = TRUE, strip.position = "top",
seed = sample(2000:3000, 1)) {
facet <- ggplot2::facet_wrap(~.bootstrap, nrow = nrow, ncol = ncol, scales = scales,
shrink = shrink, strip.position = strip.position)
facet$params$n <- n_facets
facet$params$prop <- prop
facet$params$seed <- seed
ggplot2::ggproto(NULL, FacetBootstrap,
shrink = shrink,
params = facet$params
)
}
map_data_bootstrap <- function(data, layout, params) {
if (is.null(data) || nrow(data) == 0) {
return(cbind(data, PANEL = integer(0)))
}
n_samples <- round(nrow(data) * params$prop)
set.seed(params$seed)
new_data <- lapply(seq_len(params$n), function(i) {
cbind(data[sample(nrow(data), n_samples, replace = T), , drop = FALSE], PANEL = i)
})
do.call(rbind, new_data)
}
FacetBootstrap <- ggplot2::ggproto("FacetBootstrap", ggplot2::FacetWrap,
compute_layout = compute_layout_sample,
map_data = map_data_bootstrap
)Test it out…
# facet_bootstrap
ggplot(data = mtcars) +
aes(x = wt) +
aes(y = mpg) +
facet_bootstrap(n_facets = 9,
nrow = 3) +
geom_count(alpha = .2) +
geom_smooth(method = lm, se = F)
#> `geom_smooth()` using formula = 'y ~ x'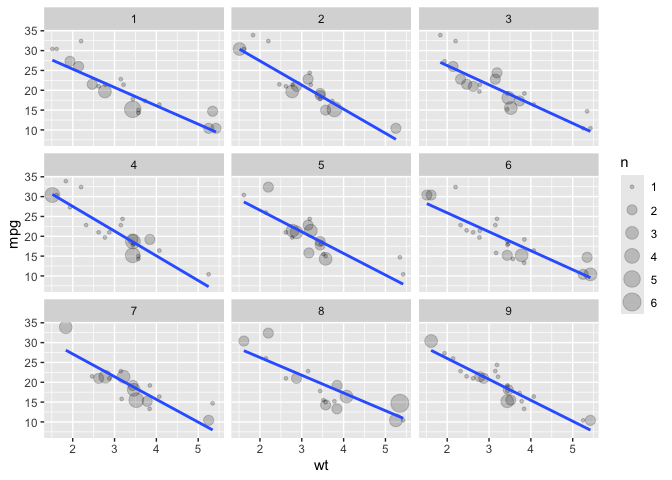
facet_scramble
#' Title
#'
#' @inheritParams facet_sample_prop
#'
#' @return
#' @export
#'
#' @examples
#' library(ggplot2)
#' set.seed(232)
#' ggplot(data = cars) +
#' aes(x = speed) +
#' aes(y = dist) +
#' geom_rug() +
#' geom_point(color = "springgreen4",
#' alpha = .75) +
#' ggxmean:::geom_corrlabel() +
#' labs(subtitle = "Pearson Correlation Coefficient(s)") +
#' facet_scramble(n_facets = 1) +
#' labs(title = "Disassociating variables") +
#' facet_scramble(n_facets = 2) +
#' facet_scramble(n_facets = 3) +
#' facet_scramble(n_facets = 12) +
#' ggxmean::geom_lm()
#'
facet_scramble <- function(n_facets = 9, prop = 1, nrow = NULL, ncol = NULL,
scales = "fixed", shrink = TRUE, strip.position = "top",
seed = sample(2000:3000, 1)) {
facet <- ggplot2::facet_wrap(~.bootstrap, nrow = nrow, ncol = ncol, scales = scales,
shrink = shrink, strip.position = strip.position)
facet$params$n <- n_facets
facet$params$seed <- seed
facet$params$prop <- prop
ggplot2::ggproto(NULL, FacetScramble,
shrink = shrink,
params = facet$params
)
}
shuffle <- function(dat){
for(i in 1:ncol(dat)){
dat[,i] = sample(dat[,i])
}
dat
}
map_data_shuffle <- function(data, layout, params) {
if (is.null(data) || nrow(data) == 0) {
return(cbind(data, PANEL = integer(0)))
}
set.seed(params$seed)
# n_samples <- round(nrow(data) * params$prop)
new_data <- lapply(seq_len(params$n), function(i) {
# data$y <- sample(data$y)
cbind(shuffle(data), PANEL = i)
})
do.call(rbind, new_data)
}
FacetScramble <- ggplot2::ggproto("FacetScramble", ggplot2::FacetWrap,
compute_layout = compute_layout_sample,
map_data = map_data_shuffle
)try it
# facet_scramble
ggplot(data = mtcars) +
aes(x = wt) +
aes(y = mpg) +
facet_scramble(n_facets = 9,
nrow = 3) +
geom_point(alpha = .2) +
geom_smooth(method = lm, se = F)
#> `geom_smooth()` using formula = 'y ~ x'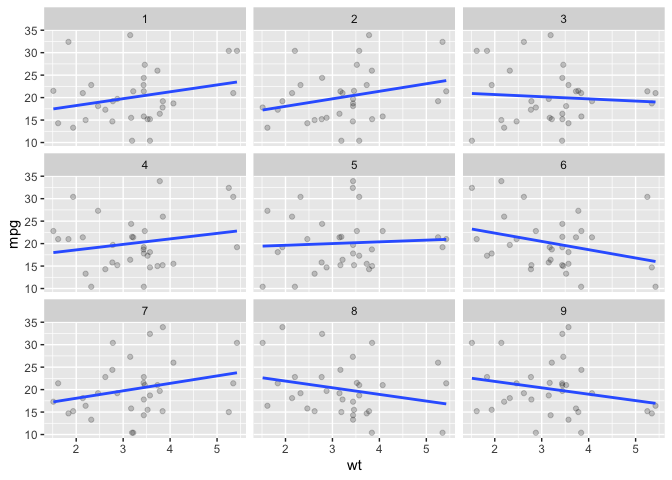
Send to R dir
library(tidyverse)
#> ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
#> ✔ dplyr 1.1.4 ✔ readr 2.1.5
#> ✔ forcats 1.0.0 ✔ stringr 1.5.1
#> ✔ lubridate 1.9.3 ✔ tibble 3.2.1
#> ✔ purrr 1.0.2 ✔ tidyr 1.3.1
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
#> ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
knitrExtra:::chunk_to_r("a_compute_layout_sample")
#> It seems you are currently knitting a Rmd/Qmd file. The parsing of the file will be done in a new R session.
knitrExtra:::chunk_to_r("facet_sample")
#> It seems you are currently knitting a Rmd/Qmd file. The parsing of the file will be done in a new R session.
knitrExtra:::chunk_to_r("facet_sample_prop")
#> It seems you are currently knitting a Rmd/Qmd file. The parsing of the file will be done in a new R session.
knitrExtra:::chunk_to_r("facet_bootstrap")
#> It seems you are currently knitting a Rmd/Qmd file. The parsing of the file will be done in a new R session.
knitrExtra:::chunk_to_r("facet_scramble")
#> It seems you are currently knitting a Rmd/Qmd file. The parsing of the file will be done in a new R session.