Proposing the {ggtemp} package! 🦄
The goal of {ggtemp} is to make writing some quick, useful extension functions succinctly (right now writing new stat_* and geom_* layers are the focus.
Currently, the amount of code required to write some extensions is a bit of a mouthful, and could feel prohibitive for day-to-day analysis scripts. Specifically, defining new geom_* and stat_* layers outside of the context of a package, I believe, is not common, but could be quite useful, ultimately making plot builds themselves intuitive and fun, and code more readable. However the usual amount of code required to make define new geom_* or stat_* functions, might feel like it ‘gunks up’ your script currently.
With the {ggtemp} package, we’ll live in a different world (🦄 🦄 🦄) where the task is a snap 🫰, and the readability of the in-script definition of a geom_* or stat_* function is quite succinct:
Proposed API where we create a new geom_* layer function.
Related:
- First experiment sketch
- Elio C’s Stat Rasa (June Choe refered me to this cool post!)
- I enjoyed working with StatRasa in my sketchbook
- ggplot2 extension cookbook might serve as reference for long-form syntax.
Issues and where this all might be going:
There doesn’t seem to be a complete match of the full functionality of the layers. For example, you have to write mapping = aes() to specify the aesthetic mapping within the layer.
- Maybe we can fix this.
- Maybe the temp utilities can be used for prototyping, and then we return to the more conventional syntax
library(ggtemp)
# 1. work out some compute
compute_group_xmean <- function(data, scales){
data |> # a dataframe with vars x, the required aesthetic
summarize(x = mean(x)) |>
mutate(xend = x) |>
mutate(y = -Inf, yend = Inf)
}
# 2. create layer function based on compute geom_xmean)
create_layer_temp(fun_name = "geom_xmean",
compute_group = compute_group_xmean,
required_aes = "x",
geom_defaut = "segment")
# 3. Use temp layer!
ggplot(data = cars) +
aes(x = speed, y = dist) +
geom_point() +
geom_xmean()Intro Thoughts
What if you just want to define a basic computational engine (geom_* or stat_* function) on the fly in a script. It seems like it requires a good amount of code, but there are things that repeat. Below, we see if we define a StatTemp within a function, and use that function to remove some of the repetition for vanilla-y extensions.
TLDR: This seems to work, and surprisingly well (??). I thought I’d only be able to use StatTemp once, but you seem to be able to define multiple geoms_* functions with the same define_temp_geom wrapper…
Status Quo: 1. compute, 2. ggproto, 3. define layer
library(tidyverse)
compute_panel_equilateral <- function(data, scales, n = 15){
data |>
mutate(group = row_number()) |>
crossing(tibble(z = 0:n)) |>
mutate(around = 2*pi*z/max(z)) |>
mutate(x = x0 + cos(around)*r,
y = y0 + sin(around)*r)
}
StatCircle <- ggproto(
`_class` = "StatCircle",
`_inherit` = ggplot2::Stat,
compute_panel = compute_panel_equilateral,
required_aes = c("x0", "y0", "r"))
geom_circle <- function(
mapping = NULL,
data = NULL,
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE, ...) {
ggplot2::layer(
stat = StatCircle, # proto object from Step 2
geom = ggplot2::GeomPolygon, # inherit other behavior
data = data,
mapping = mapping,
position = position,
show.legend = show.legend,
inherit.aes = inherit.aes,
params = list(na.rm = na.rm, ...)
)
}
data.frame(x0 = 0:1, y0 = 0:1, r = 1:2/3) |>
ggplot() +
aes(x0 = x0, y0 = y0, r = r) +
geom_circle() +
aes(fill = r)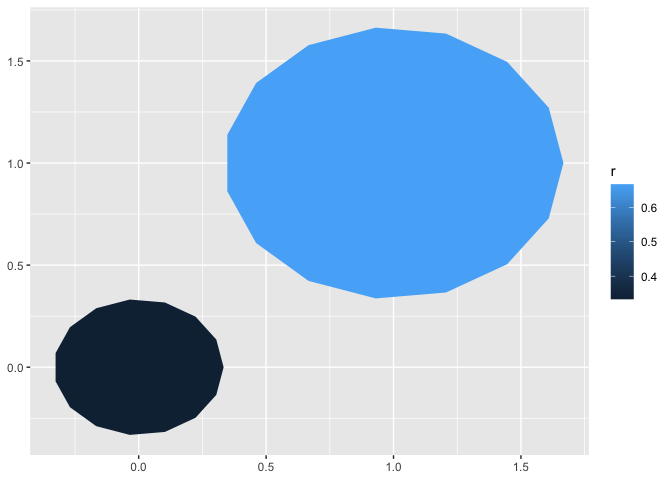
Experimental: define_layer_temp() combines 2 and 3 in using a temp Stat under the hood
`%||%` <- ggplot2:::`%||%`
define_layer_temp <- function(
default_aes = ggplot2::aes(),
required_aes = character(),
dropped_aes = character(),
optional_aes = character(),
non_missing_aes = character(),
compute_group = NULL,
compute_panel = NULL,
compute_layer = NULL,
setup_data = NULL,
# finish_layer = # we'll work on making these stat ggproto slots accessible too
# retransform
# extra_params =
# setup_params
# parameters
geom = NULL,
geom_default = ggplot2::GeomPoint,
mapping = NULL,
data = NULL,
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
...) {
StatTemp <- ggproto(
`_class` = "StatTemp",
`_inherit` = ggplot2::Stat,
default_aes = default_aes,
required_aes = required_aes)
if(!is.null(compute_group)){StatTemp$compute_group <- compute_group}
if(!is.null(compute_panel)){StatTemp$compute_panel <- compute_panel}
if(!is.null(compute_layer)){StatTemp$compute_layer <- compute_layer}
if(!is.null(setup_data)){StatTemp$setup_data <- setup_data}
if(is.null(geom)){geom <- geom_default}
ggplot2::layer(
stat = StatTemp,
geom = geom,
data = data,
mapping = mapping,
position = position,
show.legend = show.legend,
inherit.aes = inherit.aes,
params = list(na.rm = na.rm, ...)
)
}Try it out
abbreviated definition geom_circle() using define_layer_temp
compute_panel_circle <- function(data, scales, n = 15){
data |>
mutate(group = row_number()) |>
crossing(tibble(z = 0:n)) |>
mutate(around = 2*pi*z/max(z)) |>
mutate(x = x0 + cos(around)*r,
y = y0 + sin(around)*r)
}
geom_circle <- function(...){
define_layer_temp(
required_aes = c("x0", "y0", "r"),
compute_panel = compute_panel_circle,
geom_default = ggplot2::GeomPath,
...)
}use geom_circle()
We see that the layers that are created can always have there geom switched (provided that required aes are computed in the background).
library(ggplot2)
data.frame(x0 = 0:1, y0 = 0:1, r = 1:2/3) |>
ggplot() +
aes(x0 = x0, y0 = y0, r = r) +
geom_circle() +
aes(fill = r)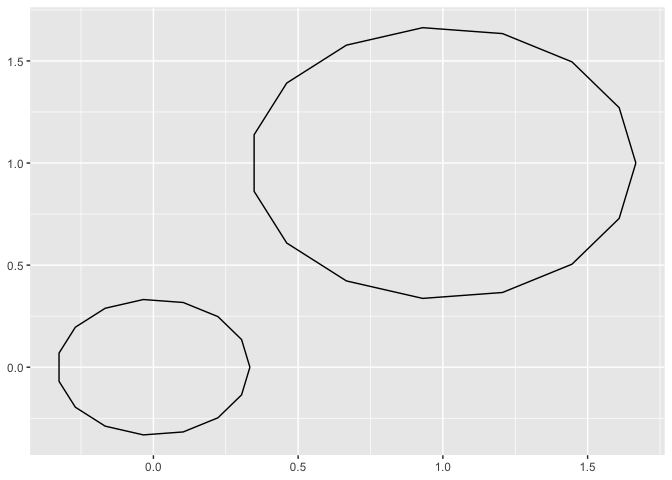
last_plot() +
geom_circle(geom = "point")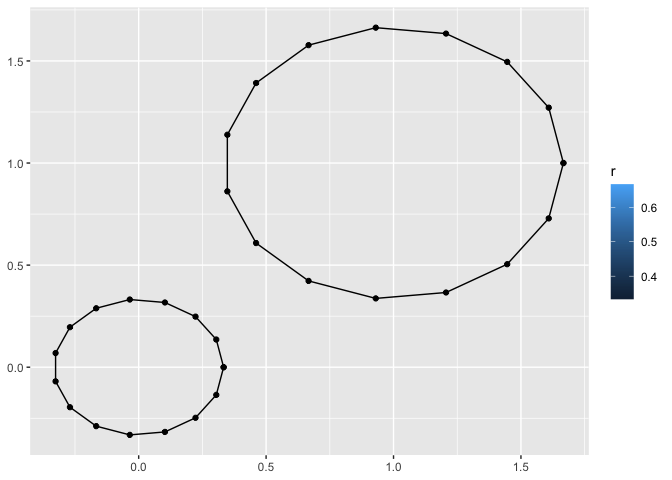
Can you define a second w/ the same StatTemp…
define geom_heart
compute_panel_heart <- function(data, scales){
data %>%
mutate(group = row_number()) %>%
tidyr::crossing(around = 0:15/15) %>%
dplyr::mutate(
y = y0 + r * (
.85 * cos(2*pi*around)
- .35 * cos(2 * 2*pi*around)
- .25 * cos(3 * 2*pi*around)
- .05 * cos(4 * 2*pi*around)
),
x = x0 + r * (sin(2*pi*around)^3))
}
geom_heart <- function(...){
define_layer_temp(
required_aes = c("x0", "y0", "r"),
compute_panel = compute_panel_heart,
geom_default =ggplot2::GeomPolygon,
...)
}try using both geom_heart and geom_circle together…
data.frame(x0 = 0:1, y0 = 0:1, r = 1:2/3) |>
ggplot() +
aes(x0 = x0, y0 = y0, r = r) +
geom_heart(alpha = .3) +
geom_circle(color = "red",
data = data.frame(x0 = 4,y0 = 2, r = 1)) +
annotate(geom = "point", x = .5, y = .5, size = 8, color = "green")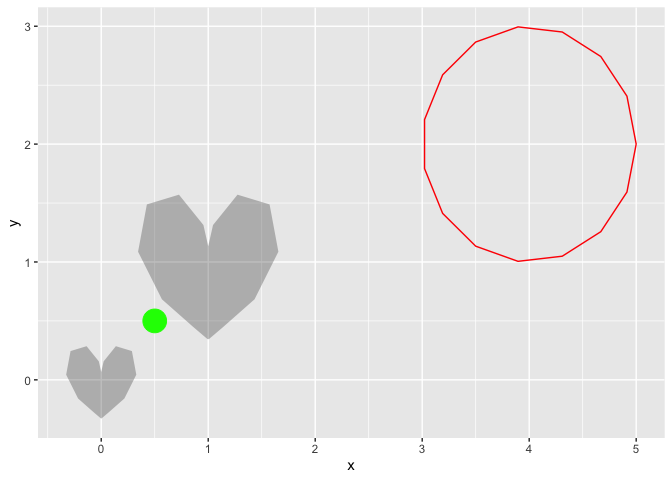
And create_layer_temp method, even more experimental (but feeling nicer to use)
wrapping this…
create_layer_temp <- function(fun_name ="geom_circle",
compute_panel = NULL,
compute_group = NULL,
required_aes = character(),
default_aes = aes(),
geom_default ="point", ...){
assign(x = fun_name,
value = function(...){
define_layer_temp(
required_aes = required_aes,
default_aes = default_aes,
compute_panel = compute_panel,
compute_group = compute_group,
geom_default = geom_default,
...) },
pos = 1
)
}Let’s do star example!
compute_panel_star <- function(data, scales, n_points = 5, prop_inner_r){
n_vertices <- n_points * 2
data %>%
mutate(group = row_number()) %>%
tidyr::crossing(around = 2*pi*0:(n_vertices)/(n_vertices)+pi/2) %>%
dplyr::mutate(
y = y + (r - r*c(rep(c(0,.35), 5), 0)
) * sin(around) ,
x = x + (r - r*c(rep(c(0,.35), 5), 0)
) * cos(around)
)
}
create_layer_temp(fun_name = "geom_star",
compute_panel = compute_panel_star,
required_aes = c("x", "y", "r"),
geom_default ="polygon")
library(ggplot2)
ggplot(cars[1:8,] ) +
aes(x = speed, y = dist, r = 1) +
geom_star() +
coord_equal()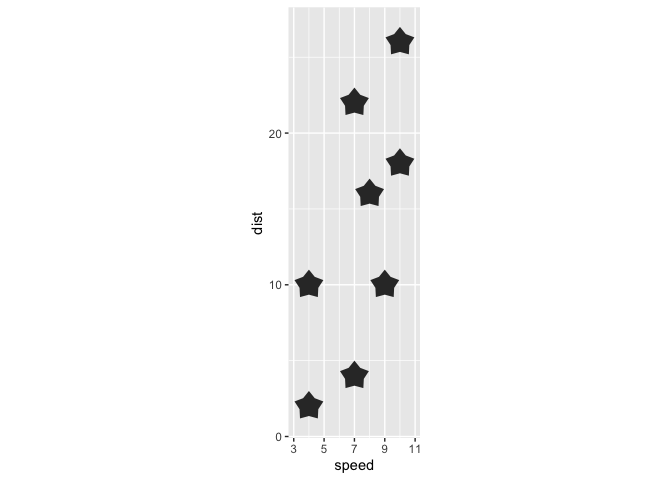
last_plot() +
geom_star(geom = "point", color = "magenta")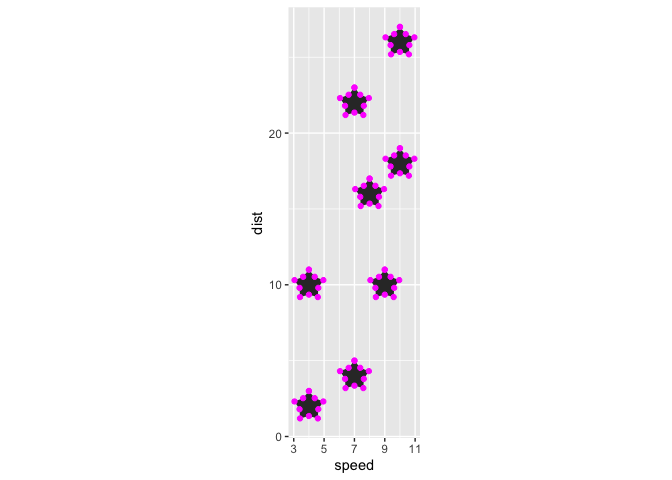
A point with no required aes
compute_group_point <- function(data, scales){
if(is.null(data$y)){data$y <- 0}
if(is.null(data$x)){data$x <- 0}
data
}
cars |>
select(x = speed, y = dist) |>
compute_group_point()
#> x y
#> 1 4 2
#> 2 4 10
#> 3 7 4
#> 4 7 22
#> 5 8 16
#> 6 9 10
#> 7 10 18
#> 8 10 26
#> 9 10 34
#> 10 11 17
#> 11 11 28
#> 12 12 14
#> 13 12 20
#> 14 12 24
#> 15 12 28
#> 16 13 26
#> 17 13 34
#> 18 13 34
#> 19 13 46
#> 20 14 26
#> 21 14 36
#> 22 14 60
#> 23 14 80
#> 24 15 20
#> 25 15 26
#> 26 15 54
#> 27 16 32
#> 28 16 40
#> 29 17 32
#> 30 17 40
#> 31 17 50
#> 32 18 42
#> 33 18 56
#> 34 18 76
#> 35 18 84
#> 36 19 36
#> 37 19 46
#> 38 19 68
#> 39 20 32
#> 40 20 48
#> 41 20 52
#> 42 20 56
#> 43 20 64
#> 44 22 66
#> 45 23 54
#> 46 24 70
#> 47 24 92
#> 48 24 93
#> 49 24 120
#> 50 25 85
create_layer_temp(fun_name = "geom_point2",
compute_group = compute_group_point,
required_aes = character(),
default_aes = aes(x = NULL, y = NULL),
geom_default = "point")
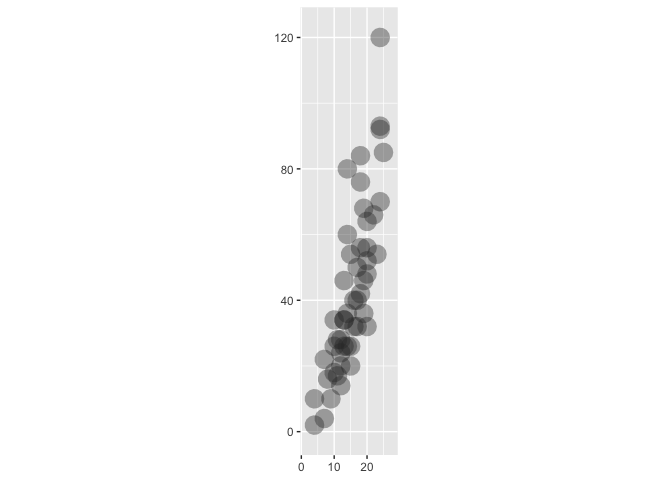
ggplot(cars) +
geom_point2(alpha = .7)
geom_xmean on the fly with compute group…
# 1. write some compute
compute_group_xmean <- function(data, scales){
data |>
summarize(x = mean(x)) |>
mutate(xend = x) |>
mutate(y = -Inf, yend = Inf)
}
# 2. define function
create_layer_temp(fun_name = "geom_xmean",
compute_group = compute_group_xmean,
required_aes = "x",
geom_default ="segment")
# 3. use function
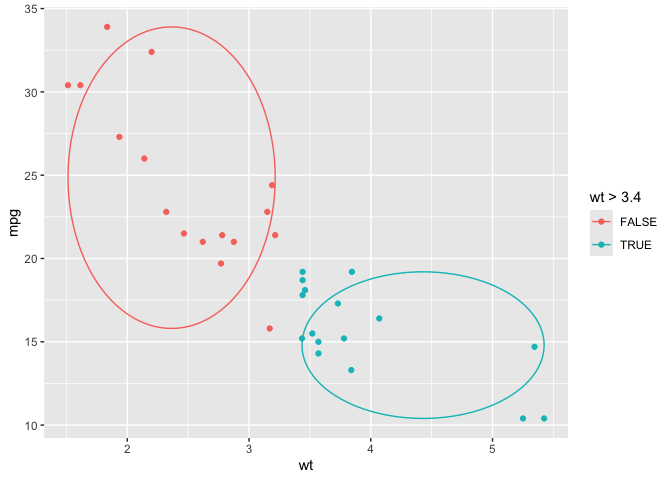
ggplot(cars) +
aes(x = speed, y = dist) +
geom_point() +
geom_xmean() +
aes(color = speed > 18)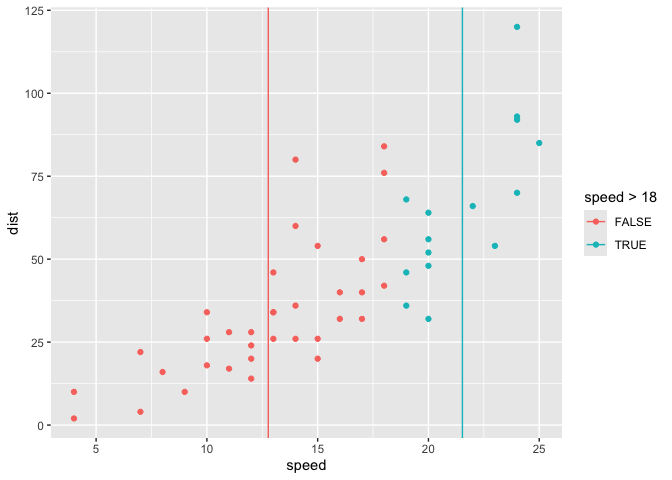
compute_oval_minmax
compute_oval_minmax <- function(data, scales, n = 100){
data |>
summarize(
x0 = sum(range(x))/2,
y0 = sum(range(y))/2,
rx = (range(x)[2] - range(x)[1])/2 ,
ry = (range(y)[2] - range(y)[1])/2) |>
# mutate(group = row_number()) |>
crossing(tibble(z = 0:n)) |>
mutate(around = 2*pi*z/max(z)) |>
mutate(x = x0 + cos(around)*rx,
y = y0 + sin(around)*ry)
}
mtcars |>
select(x = wt, y = mpg) |>
compute_oval_minmax()
#> # A tibble: 101 × 8
#> x0 y0 rx ry z around x y
#> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
#> 1 3.47 22.2 1.96 11.8 0 0 5.42 22.2
#> 2 3.47 22.2 1.96 11.8 1 0.0628 5.42 22.9
#> 3 3.47 22.2 1.96 11.8 2 0.126 5.41 23.6
#> 4 3.47 22.2 1.96 11.8 3 0.188 5.39 24.4
#> 5 3.47 22.2 1.96 11.8 4 0.251 5.36 25.1
#> 6 3.47 22.2 1.96 11.8 5 0.314 5.33 25.8
#> 7 3.47 22.2 1.96 11.8 6 0.377 5.29 26.5
#> 8 3.47 22.2 1.96 11.8 7 0.440 5.24 27.2
#> 9 3.47 22.2 1.96 11.8 8 0.503 5.18 27.8
#> 10 3.47 22.2 1.96 11.8 9 0.565 5.12 28.4
#> # ℹ 91 more rows
# 2. define function
create_layer_temp(fun_name = "geom_oval_xy_range",
compute_group = compute_oval_minmax,
required_aes = c("x","y"),
geom_default = "path")
ggplot(mtcars) +
aes(x = wt, y = mpg) +
geom_point() +
geom_oval_xy_range()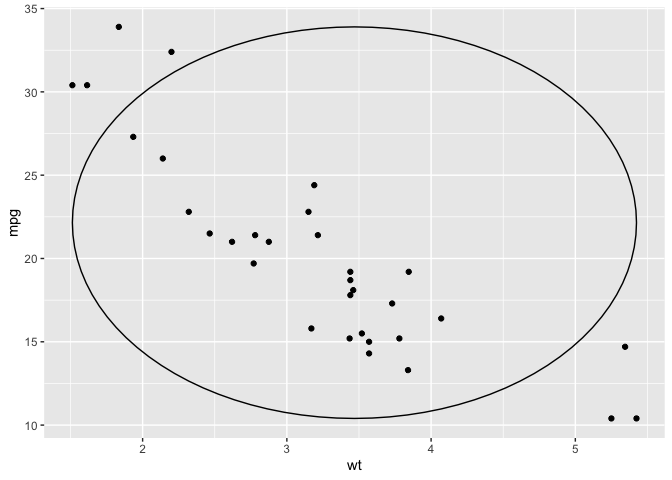
compute_group_progression <- function(data, scales){
data |>
mutate(xend = lead(x)) |>
mutate(yend = lead(y))
}
create_layer_temp(fun_name = "stat_progression",
compute_group = compute_group_progression,
required_aes = c("x","y"),
geom_default = "segment")
geom_progression <- function(...){
stat_progression(arrow = arrow(ends = "last", length = unit(.1, "in")),...)
}
tibble::tribble(~event, ~date,
"Announcement", 0,
"deadline", 3,
"extended\ndeadline", 5) |>
ggplot() +
aes(x = date, y = "Conf") +
geom_progression() +
geom_point() +
geom_text(aes(label = event), vjust = 0)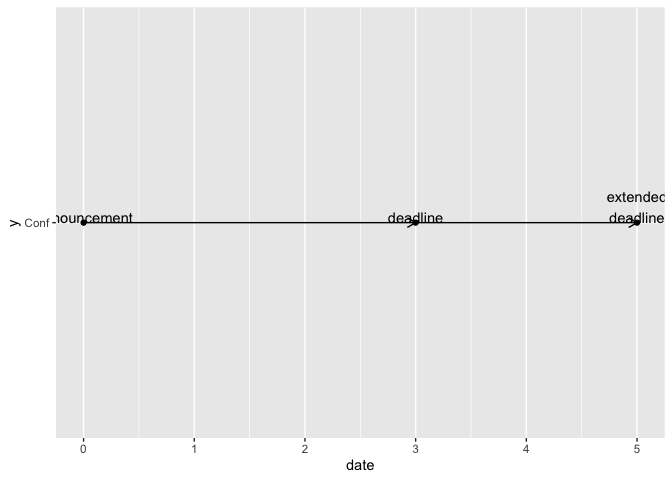
# And w/ stackoverflow example.
# https://stackoverflow.com/questions/70249589/how-to-add-multiple-arrows-to-a-path-according-to-line-direction-using-ggplot2
data.frame(long = c(0.596, 0.641, 0.695, 0.741, 0.788, 0.837,
0.887, 0.937, 0.993, 0.984, 0.934, 0.886,
0.838, 0.778, 0.738, 0.681, 0.642, 0.593),
lat = c(23.630, 24.085, 24.643, 25.067, 25.491, 25.899,
26.305, 26.670, 27.049, 27.025, 26.836, 26.636,
26.429, 26.152, 25.965, 25.664, 25.442, 24.510)) %>%
ggplot() +
aes(x = long, y = lat) +
geom_progression()
prop
setup_data_prop <- function(data, params){
data |>
mutate(x = paste("test", x))
}
compute_panel_prop <- function(data, scales, true_case = "Yes"){
if(is.null(data$weight)){data$weight <- 1}
data |>
group_by(x) |>
summarise(y = sum(weight)) |>
mutate(x_num_lab = as.numeric(x == true_case)) |>
mutate(x = as.numeric(x)) |>
mutate(x = factor(paste(x, x_num_lab)) )
}
# Titanic |>
# data.frame() |>
# summarise(weight = Freq, x = Survived) |>
# compute_panel_prop()
#
#
# create_layer_temp("geom_prop",
# setup_data = setup_data_prop,
# compute_panel = compute_panel_prop,
# geom = "col")
#
# Titanic |>
# data.frame() |>
# ggplot() +
# aes(x = Survived, weight = Freq) +
# geom_prop()spatial ‘status quo’ of ggplot2 extension cookbook
northcarolina_county_reference0 <-
sf::st_read(system.file("shape/nc.shp", package="sf")) |>
dplyr::rename(county_name = NAME,
fips = FIPS) |>
dplyr::select(county_name, fips, geometry)
#> Reading layer `nc' from data source
#> `/Library/Frameworks/R.framework/Versions/4.2/Resources/library/sf/shape/nc.shp'
#> using driver `ESRI Shapefile'
#> Simple feature collection with 100 features and 14 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
#> Geodetic CRS: NAD27
return_st_bbox_df <- function(sf_df){
bb <- sf::st_bbox(sf_df)
data.frame(xmin = bb[1], ymin = bb[2],
xmax = bb[3], ymax = bb[4])
}
add_xy_coords <- function(geo_df){
geo_df |>
dplyr::pull(geometry) |>
sf::st_zm() |>
sf::st_point_on_surface() ->
points_sf
the_coords <- do.call(rbind, sf::st_geometry(points_sf)) %>%
tibble::as_tibble() %>% setNames(c("x","y"))
cbind(geo_df, the_coords)
}
northcarolina_county_reference <- northcarolina_county_reference0 |>
dplyr::mutate(bb =
purrr::map(geometry,
return_st_bbox_df)) |>
tidyr::unnest(bb) |>
data.frame() |>
add_xy_coords()
compute_panel_county <- function(data, scales){
data |>
dplyr::inner_join(northcarolina_county_reference)
}
StatNcfips <- ggplot2::ggproto(`_class` = "StatNcfips",
`_inherit` = ggplot2::Stat,
required_aes = "fips|county_name",
compute_panel = compute_panel_county)
stat_county <- function(
mapping = NULL,
data = NULL,
geom = ggplot2::GeomSf,
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
crs = "NAD27", # "NAD27", 5070, "WGS84", "NAD83", 4326 , 3857
...) {
c(ggplot2::layer_sf(
stat = StatNcfips, # proto object from step 2
geom = geom, # inherit other behavior
data = data,
mapping = mapping,
position = position,
show.legend = show.legend,
inherit.aes = inherit.aes,
params = rlang::list2(na.rm = na.rm, ...)
),
coord_sf(crs = crs,
default_crs = sf::st_crs(crs),
datum = crs,
default = TRUE)
)
}
ggnorthcarolina::northcarolina_county_flat |>
ggplot() +
aes(fips = fips) +
stat_county(crs = "NAD83") +
aes(fill = SID74/BIR74) +
stat_county(geom = 'text',
aes(label = SID74),
color = "oldlace")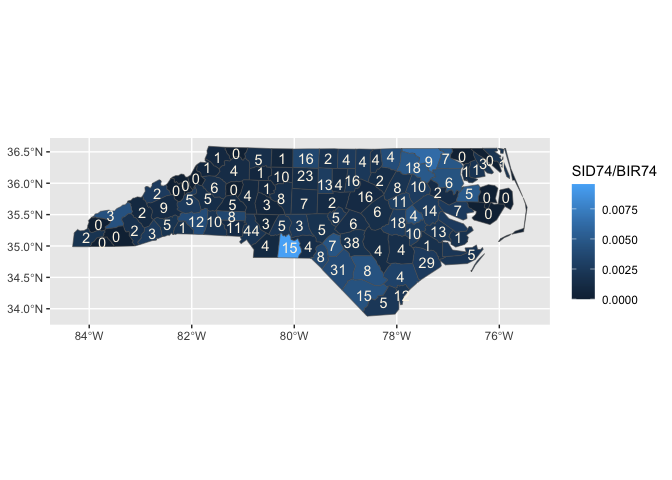
define_layer_sf_temp build
northcarolina_county_reference0 <-
sf::st_read(system.file("shape/nc.shp", package="sf")) |>
dplyr::rename(county_name = NAME,
fips = FIPS) |>
dplyr::select(county_name, fips, geometry)
#> Reading layer `nc' from data source
#> `/Library/Frameworks/R.framework/Versions/4.2/Resources/library/sf/shape/nc.shp'
#> using driver `ESRI Shapefile'
#> Simple feature collection with 100 features and 14 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
#> Geodetic CRS: NAD27
return_st_bbox_df <- function(sf_df){
bb <- sf::st_bbox(sf_df)
data.frame(xmin = bb[1], ymin = bb[2],
xmax = bb[3], ymax = bb[4])
}
add_xy_coords <- function(geo_df){
geo_df |>
dplyr::pull(geometry) |>
sf::st_zm() |>
sf::st_point_on_surface() ->
points_sf
the_coords <- do.call(rbind, sf::st_geometry(points_sf)) %>%
tibble::as_tibble() %>% setNames(c("x","y"))
cbind(geo_df, the_coords)
}
define_layer_sf_temp <- function(ref_df,
geom = NULL,
geom_default = ggplot2::GeomSf,
required_aes,
default_aes = ggplot2::aes(),
stamp = FALSE,
mapping = NULL,
data = NULL,
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
crs = sf::st_crs(ref_df),
...){
ref_df_w_bb_and_xy_centers <-
ref_df |>
dplyr::mutate(bb =
purrr::map(geometry,
return_st_bbox_df)) |>
tidyr::unnest(bb) |>
data.frame() |>
add_xy_coords()
ref_df_w_bb_and_xy_centers$id_col <- ref_df_w_bb_and_xy_centers[,1]
compute_panel_geo <- function(data, scales, keep = NULL, drop = c()){
if(!stamp){
out <- data |>
dplyr::inner_join(ref_df_w_bb_and_xy_centers) |>
filter(!(id_col %in% drop))
}
if(stamp){
out <- ref_df_w_bb_and_xy_centers |>
filter(!(id_col %in% drop))
}
out
}
StatTempsf <- ggplot2::ggproto(`_class` = "StatTempsf",
`_inherit` = ggplot2::Stat,
required_aes = required_aes,
compute_panel = compute_panel_geo,
default_aes = default_aes)
if(is.null(geom)){geom <- geom_default}
c(ggplot2::layer_sf(
stat = StatTempsf, # proto object from step 2
geom = geom, # inherit other behavior
data = data,
mapping = mapping,
position = position,
show.legend = show.legend,
inherit.aes = inherit.aes,
params = rlang::list2(na.rm = na.rm, ...)
),
coord_sf(crs = crs,
default_crs = sf::st_crs(crs),
datum = crs,
default = TRUE)
)
}Try it out
sf::st_read(system.file("shape/nc.shp", package="sf")) |>
dplyr::rename(county_name = NAME,
fips = FIPS) |>
dplyr::select(county_name, fips, geometry) ->
nc_reference
#> Reading layer `nc' from data source
#> `/Library/Frameworks/R.framework/Versions/4.2/Resources/library/sf/shape/nc.shp'
#> using driver `ESRI Shapefile'
#> Simple feature collection with 100 features and 14 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
#> Geodetic CRS: NAD27
geom_county2 <- function(...){
define_layer_sf_temp(ref_df = nc_reference,
required_aes = "fips|county_name",
default_aes = aes(label = after_stat(county_name)),
...)
}
ggnorthcarolina::northcarolina_county_flat |>
ggplot() +
aes(fips = fips) +
geom_county2() +
aes(fill = SID74/BIR74) +
geom_county2(geom = "text",
color = "pink")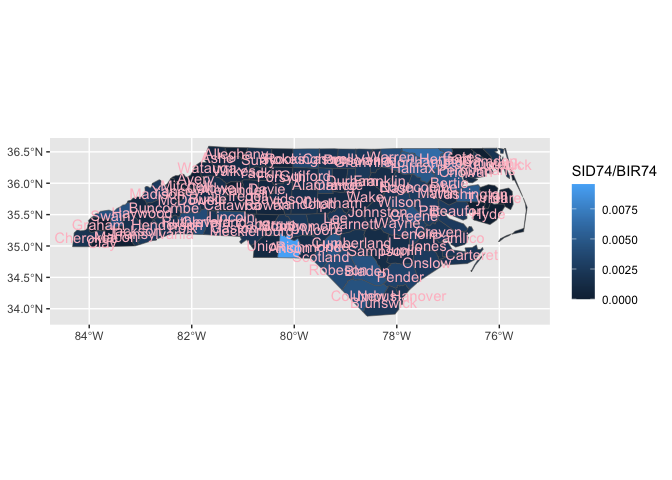
create_layer_sf_temp <- function(ref_df,
fun_name ="geom_my_sf",
required_aes,
default_aes = ggplot2::aes(),
geom_default = ggplot2::GeomSf,
...){
assign(x = fun_name,
value = function(...){
define_layer_sf_temp(ref_df = ref_df,
required_aes = required_aes,
geom_default = geom_default,
default_aes = default_aes,
...) },
pos = 1
)
}
sf::st_read(system.file("shape/nc.shp", package="sf")) |>
dplyr::rename(county_name = NAME,
fips = FIPS) |>
dplyr::select(county_name, fips, geometry) ->
my_ref_data
#> Reading layer `nc' from data source
#> `/Library/Frameworks/R.framework/Versions/4.2/Resources/library/sf/shape/nc.shp'
#> using driver `ESRI Shapefile'
#> Simple feature collection with 100 features and 14 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
#> Geodetic CRS: NAD27
create_layer_sf_temp(ref_df = my_ref_data,
fun_name = "geom_county",
required_aes = "county_name|fips",
default_aes = ggplot2::aes(label = after_stat(county_name)))
ggnorthcarolina::northcarolina_county_flat |>
ggplot() +
aes(fips = fips) +
geom_county() +
aes(fill = SID74/BIR74) +
geom_county(geom = "text",
mapping = aes(label = BIR74)) # oh ho!!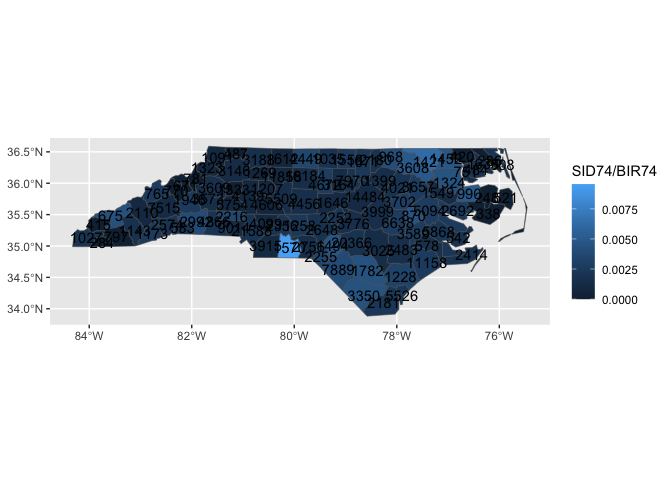
ggnorthcarolina::northcarolina_county_flat |>
ggplot() +
aes(fips = fips) +
geom_county() +
aes(fill = SID74/BIR74) +
geom_county(geom = "text", color = "pink",
check_overlap = T)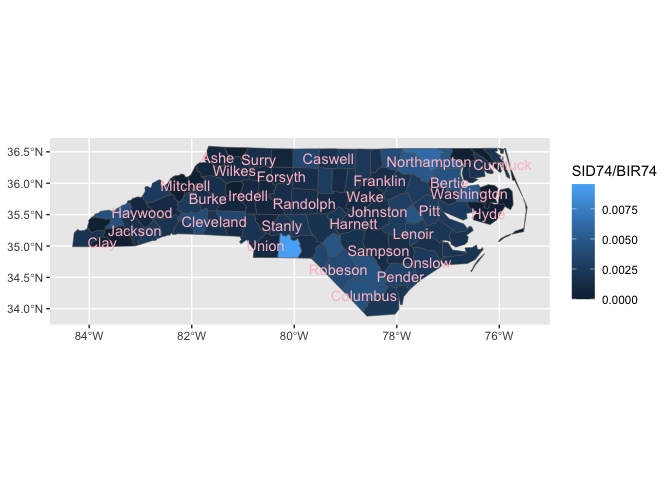
library(tmap)
data(NLD_prov)
data("NLD_muni")
# create geo reference data frame
NLD_prov |>
dplyr::select(prov_code = code, prov_name = name, geometry) ->
netherlands_prov_ref_geo
NLD_muni |>
dplyr::select(muni_code = code, muni_name = name, geometry) ->
netherlands_muni_ref_geo
# create new geom_* function
create_layer_sf_temp(ref_df = netherlands_prov_ref_geo,
fun_name = "geom_nl_prov",
required_aes = "prov_name|prov_code",
default_aes = aes(label = after_stat(prov_name)))
create_layer_sf_temp(ref_df = netherlands_muni_ref_geo,
fun_name = "geom_nl_muni",
required_aes = "muni_name|muni_code",
default_aes = aes(label = after_stat(muni_name)))
# Make a map
NLD_prov |>
sf::st_drop_geometry() |>
ggplot() +
aes(prov_code = code) +
geom_nl_prov() +
geom_nl_prov(geom = "text") +
aes(fill = pop_15_24) 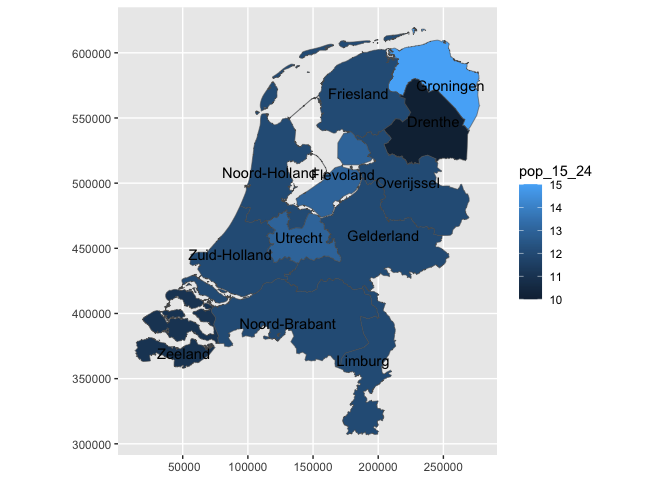
NLD_muni |>
sf::st_drop_geometry() |>
ggplot() +
aes(muni_code = code) +
geom_nl_muni() +
geom_nl_muni(geom = "text",
data = . %>% filter(population > 100000 ),
check_overlap = T,
color = "gray80",
face = "bold") +
aes(fill = pop_15_24) +
scale_fill_viridis_c() +
ggstamp::theme_void_fill("grey")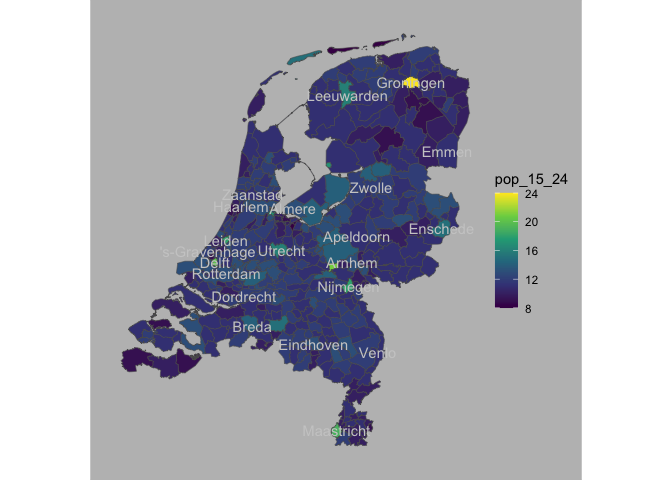
rnaturalearth::ne_countries(
scale = "medium", returnclass = "sf") %>%
select(name, continent, geometry, iso_a3) %>%
rename(country_name = name,
iso3c = iso_a3
) ->
ref_data
create_layer_sf_temp(ref_df = ref_data,
fun_name = "geom_country",
required_aes = "country_name|iso3c")
gapminder::gapminder |>
filter(year == 2002) |>
ggplot() +
aes(country_name = country) +
geom_country() +
geom_country(geom = "text",
mapping = aes(label = country),
check_overlap =T)
library(tidyverse)
heritage_wide <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2024/2024-02-06/heritage.csv')
heritage <- heritage_wide |>
pivot_longer(-1, names_to = "year", values_to = "count") |>
mutate(year = as.numeric(year)) |>
mutate(country = as.factor(country))
heritage |>
ggplot() +
aes(country_name = country) +
geom_country() +
aes(fill = count) +
facet_grid(~year) +
geom_country(geom = "text", mapping = aes(label = paste(country, count, sep = "\n")))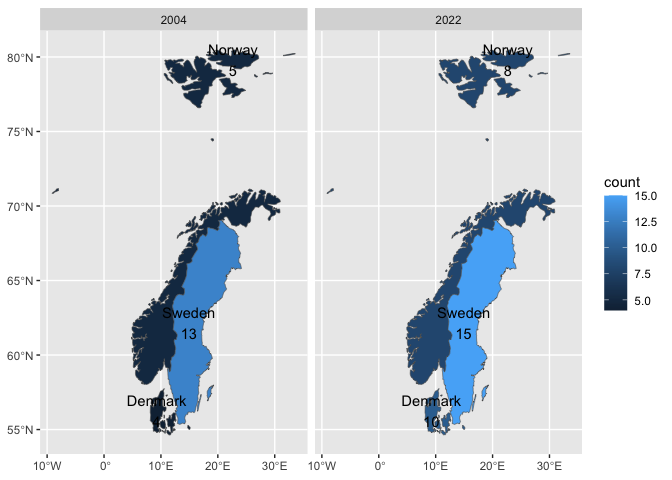
Part II. Packaging and documentation 🚧 ✅
Phase 1. Minimal working package
- Bit A. Created files for package archetecture, running
devtools::create(".")in interactive session. ✅ - Bit B. Added roxygen skeleton? 🚧
- Bit C. Managed dependencies ? 🚧
usethis::use_package("ggplot2")- Bit D. Moved functions R folder? ✅
readme2pkg::chunk_to_r("define_layer_temp")
readme2pkg::chunk_to_r("create_layer_temp")
readme2pkg::chunk_to_r("define_layer_sf_temp")
readme2pkg::chunk_to_r("create_layer_sf_temp")- Run
devtools::check()and addressed errors. 🚧
devtools::check(pkg = ".")- ✅
devtools::build()- Bit G. Write and test traditional README that uses built package. 🚧 ✅
Install package with:
Then…
library(ggtemp) ##<< change to your package name here
compute_panel_circle <- function(data, scales, n = 15){
data |>
mutate(group = row_number()) |>
crossing(tibble(z = 0:n)) |>
mutate(around = 2*pi*z/max(z)) |>
mutate(x = x0 + cos(around)*r,
y = y0 + sin(around)*r)
}
geom_circle_points <- function(...){
ggtemp:::define_layer_temp(
required_aes = c("x0", "y0", "r"),
compute_panel = compute_panel_circle,
geom_default =ggplot2::GeomPoint,
...)
}
library(ggplot2)
ggplot(cars) +
aes(x0 = speed, y0 = dist, r = 1) +
geom_circle_points()
- Bit H. Chosen a license? 🚧 ✅
usethis::use_mit_license()- Bit I. Add lifecycle badge (experimental)
usethis::use_lifecycle_badge("experimental")Phase 2: Listen & iterate 🚧 ✅
Try to get feedback from experts on API, implementation, default decisions. Is there already work that solves this problem?
Phase 3: Let things settle
Appendix: Reports, Environment
Environment
Here I just want to print the packages and the versions
all <- sessionInfo() |> print() |> capture.output()
all[11:17]
#> [1] ""
#> [2] "attached base packages:"
#> [3] "[1] stats graphics grDevices utils datasets methods base "
#> [4] ""
#> [5] "other attached packages:"
#> [6] " [1] ggtemp_0.0.0.9000 tmap_3.3-4 lubridate_1.9.2 "
#> [7] " [4] forcats_1.0.0 stringr_1.5.0 dplyr_1.1.0 "Non-developer introduction to package (and test of installed package)
The goal of the {xxx} package
To install the dev version use the following:
Example using package
library(mypackage)
myfunction(mtcars)